1. INTRODUCTION
Water is a key resource to sustain life on earth. Better access to clean water, sanitation services, and water management create tremendous strategies for the economic growth of the country. Water pollution due to various effluents is a cause of concern worldwide. There is a rising demand for adopting practices to minimize water pollution. Among all industries, the textile industry is one of the leading contributors to pollution by liquid effluents and the reason is the use of high quantities of water in the process of dyeing [1]. One of such industrial waste that is considered as hazardous pollutant is dye. Dye is an organic (natural or synthetic chemical), colored compound which has an affinity towards the substrate to which it is being applied by giving it an everlasting color. These have been applied to products such as paint [2], plastic [3], textile [4] printing ink [5], pharmaceuticals [6], cosmetics [7], photographic, and paper [8]. According to estimates, more than 10,000 dyes and pigments are put to industrial use and over 7 × 105 tons of synthetic dyes are annually produced across the world and 10% of which is released into the water [9].
Azo dyes are most extensively used in the textile industry (60–70%) [10]. These have nitrogen-nitrogen (N=N) in their chemical structure. The azo group is generally connected to naphthalene and benzene rings and could also be attached to enolizable aliphatic groups or aromatic heterocyclic. These attached side groups to the ring are necessary for the dye to impart the color, with a variety of shades and intensity of color [11]. Azo dyes and their degradation products have been proven toxic and mutagenic to aquatic animals and humans [12]. The dyeing and dye industries do not take precautionary measures as per the legislation and freely release these toxic chemicals into the environmental sink. Color is one of the obvious water pollution indicators and is usually the first recognized contaminant [13]. The dye presence in water bodies interferes with the penetration of sunlight [14] and adversely affects the aquatic ecosystem. Some of the azo dyes are mutagenic and very toxic to living organisms [15]. Their discharge in water bodies could result in serious health issues and can have chronic and acute effects on aquatic life [16].
Several factors, including the type of dye, wastewater composition, environmental fate, and operation and handling costs of generated waste products, determine the economic and technical feasibility of each single dye removal technique. Different physicochemical methods such as membrane filtration, flocculation or coagulation, ion exchange (Dotto et al., 2012), adsorption, electrolysis, activated carbon, Fenton’s reagent, ozonation, and advanced oxidation process are utilized by industries to remove dye from the effluents. Such methods are often very costly, less efficient, and inapplicable to a wide dyes variety [17]. Moreover, these methods generate concentrated sludge which creates a disposal problem [18].
Bioremediation or utilization of microbial-based techniques to deal with pollution is a key area of research in the field of environmental sciences where naturally developed or acclimatized microbes are used to transform various toxic chemical compounds into less toxic forms. In the process of biological treatment, microorganisms follow two methods: adsorption on microbial biomass and dye biodegradation by the microbial cells [19]. Adsorption of dye may be on live cells of microbes or dead cells of microbes and this type of adsorption on biomaterial is also known as biosorption, where the dye original structure remains untouched, that is, not fragmented. On the hand in biodegradation, the structure of the original dye is fragmented or broken by microorganisms, thus achieving complete mineralization in certain cases, that is, conversion to some inorganic salts, CO2 and H2O. On studying the mechanism involved in the two processes (biosorption and biodegradation), biodegradation was more convincing. The process of biosorption does not remove or destroy the dye, instead entraps the dye in biomatrix (microbial cells). Dumping of these microbial cells having absorbed dye itself is a roadblock in their projected role in the bio-cleaning of colored water [20]. Thus, because of the disposal of adsorbed biomass, biosorption might not be the pragmatic approach for treating colored effluent from industries [21]. The effectiveness of microbial decolorization depends on the activity and adaptability of the selected microorganisms. Several microbiomes, including bacteria, fungi, and algae, are capable of biodegrading azo dyes, among which bacteria represent a most promising tool for the removal of the azo dye from textile effluents [22]. The environmentally friendly and efficient technologies development to decrease the content of dye in wastewater is of utmost important. An insight into the present review detailed the role of microbial systems in the bioremediation of environmental pollutants, their mechanism, and factors affecting biodegradation.
2. CLASSIFICATION OF DYES
The society of dyers and colorists (TDC) and the American Association of Textile Chemists and Colorists had classified the dyes based on their color, structure, and application method which are revised every 3 months since 1924. A C.I. (Color index number) generic name was given to each dye based on the color as well as its application and characteristics. Dyes based on their origin are classified into two types, namely, natural and synthetic.
2.1. Natural Dye
The dyes obtained from natural sources are called natural dyes. They are mostly applied to textiles using mordants. These mordants are metallic salts that have a high affinity for fiber as well as any coloring matter. These metallic mordants after mixing with dye in fiber form an insoluble precipitate, leading to both dye and mordant getting fixed [23]. On the basis of the chemical constitution, a natural dye can further be classified namely, indigoid dyes, alpha-hydroxynaphthoquinones, flavones, dihydropyrans, anthocyanidins, and carotenoids. Indigoid dye’s most common examples are indigo and Tyrian purple [24]. This dye type is obtained from the leaves of a wood plant as it possesses indigo as the key component in dyeing. Oppositely, alpha-hydroxy-naphthoquinones is a lawsone also known as Heena. It is mainly cultivated in Egypt and India. This dye type is also found in the unripe walnuts shell which gives orange color shades [25]. Flavones are colorless organic compounds. In general, natural yellows are derivatives of methoxy and hydroxyl substituted isoflavones and flavones. Weld (containing luteolin pigment) is a common example, which gives bright and fast colors to silk and wool [26]. Dihydropyrans are similar in chemical structure to flavones. Some examples are hematin and its leuco form, hematoxylin. These categories of natural dyes give dark shades to wool, silk, and cotton. Some of the common sources of these dyes are brazilwood, sappan wood, and logwood. Another dye, anthocyanidins, naturally possesses orange dye for cotton, and these are extracted from Bignonia chica leaves. Carotene is derivative of orange pigment found in carrots. Carotenoids owe their color to the conjugated double bonds which are present in their structure. Annatto and saffron are examples of carotenoids [27].
2.2. Synthetic Dyes
Synthetic dyes are man-made petrochemicals that may consist of lead, mercury, copper, sodium chloride, toluene, chromium, and benzene. Synthetic dyes have a brighter and wider range of colors, longer color permanence, and are easier, faster, and cheaper to produce in comparison with natural dyes. There are over 10,000 different types of synthetic dyes. Synthetic dyes are categorized on the basis of their chemical composition and the method of their application in the dyeing process. Although natural dyes are important from the chemical and historical point of view, these dyes are very expensive, need purification, and do not bind well because they lack the chemical grouping required to react with the binding sites of a fabric. Therefore, for dyeing applications, synthetic colors are utilized in place of natural dyes. This is because synthetic dyes are generally purer, less expensive, and their colors bind faster to the fabric [28]. Synthetic dyes are classified into three categories including anionic, non-ionic acid, and azo dyes. Anionic synthetic dyes include acid and whereas non-ionic dyes are basic, dyes are also known as cationic. On the other hand, dyes can be classified based on their chemical structure into anthraquinone dyes and azo dyes [29].
Azo dyes are characterized by being strong, having good all-around properties, and being less expensive. This dye has at least one azo bond (-N=N-), along with one or more aromatic structures. Azo dyes are intended to transport resistance and high photolytic stability toward major oxidizing agents. They have wide variety of applications in textile, food making, and cosmetic industries [3]. After azo dyes, anthraquinone dyes are widely used textile dyes [30]. Anthraquinone dyes have a wide color range and it almost covers the whole visible light spectrum. Dyes based on anthraquinone are the most resistant to degradation because of their fused aromatic structures, which retain the color for a duration [31]. Acid dyes are soluble in water and is used for silk, wool, nylon, modified acrylics, leather, paper, ink-jet printing, cosmetics, and food. The principal chemical classes of acid dyes are azo (including premetallized), anthraquinone, azine, triphenylmethane, nitro, nitroso, and xanthene [32]. Metal complex dyes consist of dyestuff and metal (usually chrome such as chromium, cobalt, nickel, and copper) and are being used since the 1940s. On the other hand, direct dyes are compounds of poly azo having some phthalocyanines, oxazines, and stilbenes. These dyes types are the anionic dye having water-soluble properties. They are utilized in dying rayon, cotton, nylon, leather, and paper [33]. Basic dyes are utilized on polyacrylonitrile, paper, cation dyeable polyethylene terephthalate modified polyesters, modified nylons, and in medicines. Dyes yield colored cations in a solution and they are known as cationic dyes. The chemical structure of basic dyes encompasses triarylmethane, aromatic methane, xanthene, thiazine, hemicyanine, oxazine, cyanine, and acridine [32,33].
Another dye, reactive dye is mainly used for dyeing cotton, silk, wool, nylon, and leather. This class of dyes makes a covalent bond with the fiber. It is so tightly fixed that they cannot be removed even in hash conditions [34]. Sulfur dyes have a complex structure and it contains sulfur. This class of dyes is economically friendly as they are low priced and has good wash fastness properties. These dyes are not brightly colored and are used for dyeing cotton, rayon, polyamide fibers, paper, leather, silk, and wood [33]. Vat dyes are fast dyes and are used for coloring cellulosic fiber, mainly cotton fiber. These dyes belong to the chemical class comprising indigoids and anthraquinone derivatives (including polycyclic quinones) [32]. Disperse dyes are primarily used on polyester and to a lesser amount on nylon, acrylic fibers, cellulose, and cellulose acetate. They are insoluble in water and non-ionic in nature. Disperse dyes molecule is based on azobenzene or anthraquinone molecules having nitro, hydroxyl, and amine groups attached [32,33].
3. DYES: ENVIRONMENTAL CONCERN
Wastewater released from the textile industry has many substances which are polluting in nature, like organochloride-based pesticides, and heavy metals [35]. Synthetic dyes are modeled in such a manner that they are recalcitrant and resist fading on treatment with water, soap, sweat, light, or any type of oxidizing agent [18]. The textile industry is the main producer of wastewater because water is the key component in the process of dyeing [36]. It had been estimated that 100 L of water was being used for the processing of 1 kg of textile materials. Every year all around the world almost 280,000 tons of textile dyes get discharged into industrial wastewater [37]. The appearance of color in the effluent is the leading sign that water has been polluted and the release of this colored effluent affects negatively the main water stream [38]. These water streams, when utilized in agriculture, have an adverse impact on the environment which, in turn, affects the health of living organisms [39]. These colored effluents when mixed with water bodies reduce the light penetration capacity of the water body and thus have a negative impact on aquatic flora and fauna by affecting the food chain. Even at a 1 mg/L concentration some of the dyes could be observed in water [40]. Dye-containing effluent also has been shown to increase the contaminated water’s biochemical oxygen demand [41]. Out of all the known dyes, azo dyes are the major group of synthetic dyes and are extensively used [42]. Textile effluent has been characterized by the presence of undefined organic pollutants, dyes, increased COD (organic compounds), and high conductivity due to a high amount of dissolved salt, sulfide, halogen, and heavy metals [43]. Maximum dyes pose health risks to all kinds of life forms because of their non-biodegradable nature.
Azo dyes have been known as potential health hazards. Several azo dyes have also been the cause of DNA damage which leads to malignant tumors [44]. When these compounds contacted the body of humans through skin or injection and ingestion, they are metabolized inside the mammalian liver and gastrointestinal tract by the azoreductases and converted into aromatic amines and free radicals. Azo dyes are known as relatively persistent pollutants as they are not easily degraded under aerobic conditions. Under limited oxygen conditions, these dyes could be reduced by intestinal bacteria and other microbes to colorless amines. The converted amines are toxic, mutagenic, and carcinogenic to humans and animals [40,45].
4. METHODS OF REMEDIATION OF DYES
Dye could be removed by three methods namely, physical, chemical, and biological. The conventional treatment methods, that is, physical and chemical have been ineffective in dealing with effluents containing synthetic dyes which are highly unstable chemical pollutants. There have been remarkable achievements in the use of biotechnological methods in recent years. Chemical and physical methods are used for the decolorization of colored effluent and were subjective to physiochemical factors such as dye interaction, particle size, temperature, sorbent surface area, pH, and contact time [46]. On the other hand, the biological method uses microbial cells for the bioremediation of dye.
4.1. Physical Method
There are several physical methods through which dye is biodegraded such as adsorption, membrane filters, coagulation/flocculation, ion exchange, and radiation. Adsorption is an effective and attractive method for dye elimination from wastewater, particularly if the adsorbent is not expensive and commonly available [47]. Activated carbon is mainly utilized for dye removal with great success due to its higher adsorption capacity [48]. The cellular structure of peat makes it a perfect choice as an adsorbent. It can adsorb polar organic compounds and transition metals from colored effluents. Unlike activated carbon, it requires no activation and is less expensive [49,50]. Wood chips have high-quality adsorption capabilities for acid dyes, although because of their hardness and longer contact times are required [51]. Many low-priced adsorbents have been studied on fly ash for dye adsorption [52]. Its adsorption ability depends on various properties of the adsorbent: Porous chemical structure, porous structure, and surface area [53]. Silica gel could be considered effective for removing basic dyes. However, side reactions such as air binding and air fouling with particulate matter prevent it from becoming effective for commercial use [18].
Membrane filter methods have several applications by improving the finished product quality, increasing the amount of yield generation, saving raw materials, or recovery of products from waste [54]. This method has some special features which are not found in other methods, namely, resistance to temperature, an adverse microbial attack, and chemical environment [18]. Concentrated sludge production is the main disadvantage. Ion exchange resins were used to decolorize the textile wastewater and to reduce the COD [55]. Most of the dyes are either anionic or cationic in nature; therefore, it was possible to remove them through ion exchange resins. These are not utilized extensively for the treatment of colored effluents, mainly because they couldn’t accommodate a wide spectrum of dyes. The whole process is possible only in the presence of organic solvents during regeneration time [18].
Coagulation/flocculation is the only cost-effective method used for the removal of color from the effluent. The primary wastewater treatment method proceeds with coagulation and flocculation by removing colloidal particles of turbidity, color, and bacteria. Coagulants such as Fe (III) or Al (III) salts are used in high concentrations for initiating precipitation and settling along with it [56]. Generation of the huge toxic sludge and disposal amount has been the major drawback of this method. Radiation, in general, is the emission of any rays or particles from a source. Radiation is classified into two main categories: Ionizing radiation and non-ionizing [57]. The rate of reaction was monitored by radiation dose and the amount of oxygen present in the solution. It is a proven effective method for the removal of the dye at the same time toxic organic compounds. Almost 90% of the color has been removed from the effluent. The whole process of radiation is very expensive which makes it less feasible for use. Finally, the drawbacks of all such processes have been mainly due to low efficiency, high cost, problems in the disposal, and limited versatility. Thus, most of the chemical and physical methods for the treatment of colored wastewater are not widely used in textile industry sites or plants [37].
4.2. Chemical Method
In the chemical method, various types of chemical reagents and treatments are applied to degrade the dyes such as ozone treatment, Fenton’s reagent, photochemical oxidation, sodium hypochlorite (NaOCl), and electrochemical. Fenton’s reagent is an effective decolorization chemical method of the textile wastewater which is unaffected to a biological method or is toxic to living biomass but not useful in reducing its COD if not combined with one more process such as coagulation [58]. In the acidic mixture, iron (II) acts as a catalyst which when acts on hydrogen peroxide, immediately leads to the formation of hydroxide radicals. These hydroxide radicals help in decolorizing dye wastes. This method can act on both insoluble and soluble dyes. Sludge generation is the major disadvantage of this process [59]. Ozone treatment is widely applied in the treatment of water; ozone either alone or in combinations (O3-UV or O3-H2O2) is used for treating industrial effluents [60]. Chromophore groups are accountable for color which can be fragmented by ozone either directly or indirectly through the formation of smaller fragments [61]. The only disadvantage is the short shelf life (20 min). Photochemical oxidation is the traditional technique used for industrial textile effluent treatment [62]. UV light activates the degradation of H2O2 into hydroxyl radicals which may attack and release hydrogen atoms from organic molecules capable of the oxidizing organic compound. The formation of byproducts is a major drawback [39,63].
NaOCl acts on the amino group of the dye molecule by the Cl− which starts and speeds up azo bond cleavage [64]. NaOCl, when added to effluent may remove residual colors effectively. Moreover, this method is not favored much as it has been observed to lead to the generation of toxic chlorinated compounds which are dangerous for the environment as well as human health [65]. An electrochemical method in current years has been a growing interest in wastewater treatment. This type of method has been effectively functional in the detoxification of textile wastewater [66]. Electrochemical reactions are mainly used in sulfur and vat dyeing. Electrochemical oxidation leads to the generation of hypochlorite or hydroxyl radical. These electrogenerated species are used to degrade dyes present in wastewater. However, the electricity price used is high compared to the chemicals cost [18].
4.3. Biological Method
In the last few years, awareness among the scientific community about biological techniques has increased tremendously. These techniques have several benefits over conventional techniques such as low cost, environment-friendly, safe operation, and less sludge production. Bioremediation is now considered an upcoming treatment option for dye removal in diverse conditions. The bioremediation method can use natural and recombinant microorganisms to degrade toxic materials because of flexibility in operating conditions and design. Flexibility in this technique is understood by the fact that they could be employed ex situ (off-site) or in situ (on-site) and even plants can be used (phytoremediation). Biodegradation could be defined as the biologically facilitated dye molecule breakdown into several by-products by the action of a variety of enzymes. It is a process that is energy dependent [67]. Dye biodegradation results in decolorization and the breakdown of the dye molecules into smaller fragments. Diverse microbes such as bacteria, fungi, and algae are employed for the decolorization and degradation of synthetic dyes. Microbes have various capabilities for decolorizing a variety of dyes. Some groups of microorganisms have specific advantages over others in synthetic dyes biodegradation. The bioremediation of dyes effectiveness is subject to the activity and adaptability of the microbes [38].
4.3.1. Fungi
Fungi are most effective in breaking down or sometimes completing the mineralization of synthetic dyes [68]. These degradation properties are attributed to the presence of a powerful extracellular and intracellular enzyme system comprising laccase, manganese peroxidase, and lignin peroxidase, robust morphology, and various metabolic activity [69]. The mycelia of fungal species have an advantage over unicellular organisms that they solubilize the insoluble substance by enzymes. They have a high cell-to-surface ratio which helps them to have greater enzymatic and physical contact with pollutants. Extra-cellular fungal enzymes are also advantageous in tolerating high toxicants concentration [67]. Fungi initiate the process by adsorption of dye onto hyphae, followed by the breakdown of chemical bonds by enzymes [70]. However, fungi application for the removal of the dye from textile wastewater has some inherent drawbacks such as a long growth cycle and the need for nitrogen limiting conditions [71]. Gill et al. [72] reported Congo red decolorization by Pha. chrysosporium and Dichomitus squalens. Svobodová et al. [73] have reported maximum decolorization of Reactive Orange 16 up to 80% by fungal strain Irpex lacteus. Jayasinghe et al. [74] have reported Ganoderma lucidum, Pycnoporus cinnabarinus, Pleurotus pulmonarius, Stereum ostrea, and Trametes suaveolens, for their ability to decolorize Congo red (100 mg/L).
In a report, fungal species Fusarium oxysporum, Penicillium lanosum, and Ganoderma resinaceum was reported for Blue 21 dye decolorization [75]. Synthetic dye amaranth was reported to be degraded by Bjerkandera adusta [76]. White rot fungus Armillaria spp. was confirmed for degrading azo, anthraquinone, and triphenylmethane dyes. [77]. Noval species of Alternaria alternata was able to decolorize Congo red (600 mg/L) within 48 h of incubation [78]. Ganoderma spp. has also been reported to decolorize Reactive Orange 16 [79]. Chen et al. [80] reported Coriolopsis spp. for decolorizing triphenylmethane dyes. In a report, the novel fungal strain Absidia spinosa was reported for biotransformation of Cresol Red up to 65% [81]. Barapatre et al. [82] reported Aspergillus flavus for biodegradation of Malachite Green in which intermediate N-demethylated and N-oxidized metabolites were identified [Figure 1]. Fungal strain Aspergillus bombycis was reported for having the ability to degrade Reactive Red 31 [83]. Asses et al. [84] reported Aspergillus niger having the ability to decolorize Congo red (200 mg/L) within 6 days of incubation [Figure 2]. Oudemansiella canarii, the white rot fungi, was reported for degrading Congo Red dye [85]. In another report, Aspergillus terreus was reported for degrading Direct Blue-1 [86]. Krishnan et al. [87] reported Fusarium equiseti for the degradation of methylene blue dye.
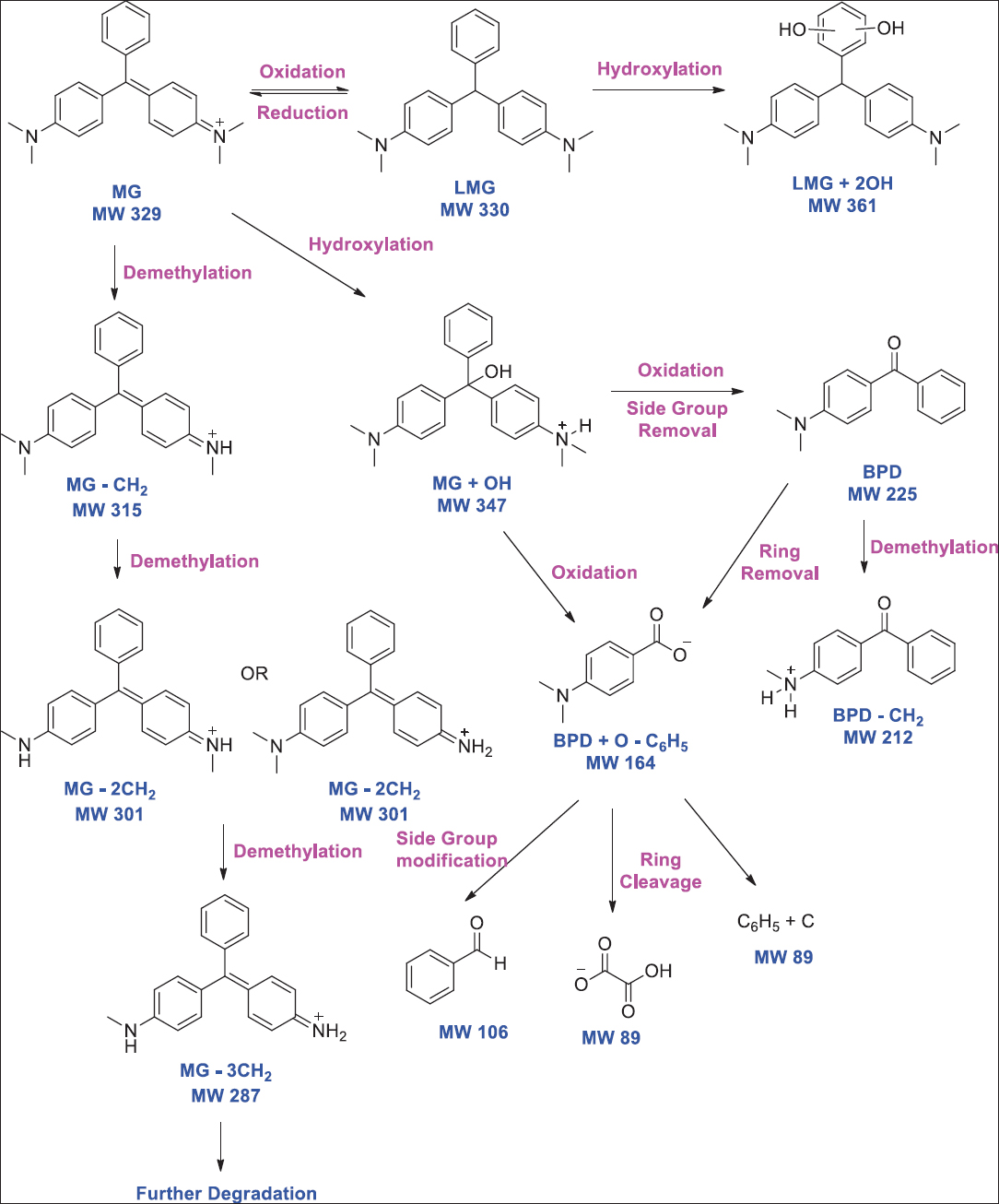 | Figure 1: Biodegradation pathway of malachite green using Aspergillus flavus. Source: Adapted with permission from Barapatre et al. [82]. [Click here to view] |
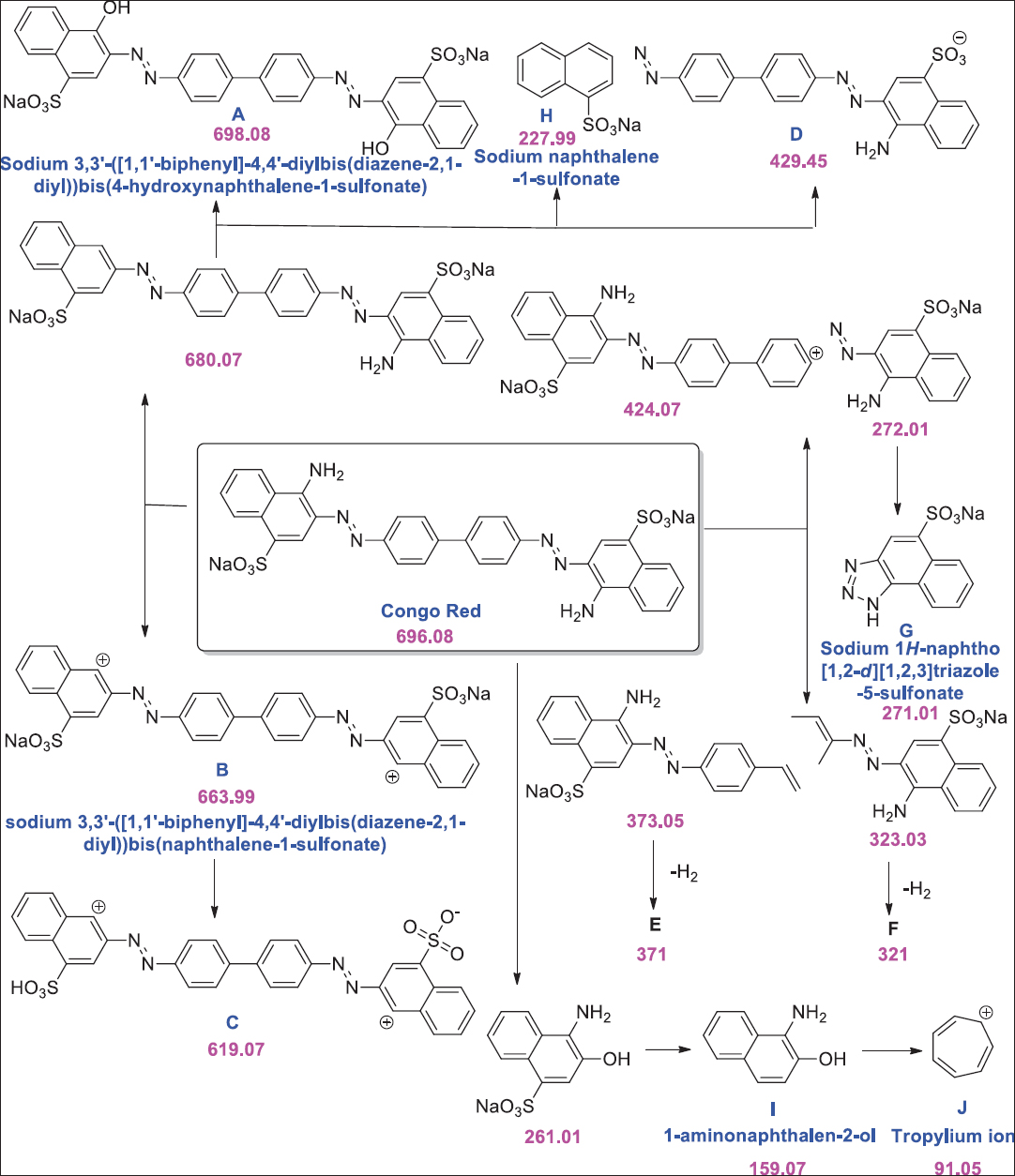 | Figure 2: Decolorization of Congo red through Aspergillus niger. Source: Adapted with permission from Asses et al. [84]. [Click here to view] |
4.3.2. Bacteria
Dyes decolorization through bacteria is faster than fungal decolorization as bacteria take less time to grow [88]. Bacterial cells represent a promising and inexpensive tool for the several azo dyes removal from the textile dye effluents. Bacteria have many advantages as compared to filamentous fungi such as higher hydraulic retention time, faster growth rate, and could be efficient in treating high strength organic wastewaters [89]. In general, the azo dyes decolorization occurs under conventional anaerobic, facultatively anaerobic, and aerobic conditions by different bacterial groups [90]. A bacteria like Pseudomonas spp. could decolorize Reactive Orange 16 (100 mg/L) to 98% within 24 h of incubation [91]. Park et al. [92] reported decolorization of Congo red (1 g/L) by Staphylococcus spp. up to 96%. In a report, Pseudomonas spp. SUK1 could decolorize methyl orange dye (300 mg/L) within 6 h of incubation [93]. On the other hand, strain Kocuria rosea exhibited complete decolorization [94]. In another study, Nocardiopsis alba could decolorize Reactive Orange 16–95% (1000 mg/L) dye within 24 h of incubation [95]. Ghanem et al. [96] reported decolorization of methyl orange (38 μg/ml 10 μg/ml, 36 μg/ml, 32 μg/ml, and 40 μg/ml) by Acinetobacter baumannii, Cytophaga columnaris, Corynebacterium spp., Escherichia coli, and Pseudomonas fluorescence up to 76, 20, 72, 64, and 80%, respectively.
A study by Shah et al. [97] showed complete decolorization of methyl orange (2.9 mg/L) by Bacillus spp. complete methyl orange decolorization has also been reported for a much higher concentration of dye (50–200 mg/L) by several researchers. In a finding by Cui et al. [98] reported 100% decolorization of Methyl orange (100 mg/L) dye by Klebsiella spp. strain Y3. Ng et al. [99] reported decolorization up to 96% for 200 mg/L of Methyl orange by Shewanella xiamenensis. Marine bacteria A. baumannii could decolorize Congo red (100 mg/L) to 99.1% within 30 h and 5 days of incubation under optimized conditions [100]. Mycobacterium could decolorize Reactive Orange 16 (250 mg/L and 100 mg/L) to 96% within 24 h [101]. Bacillus cereus, Ochrobactrum spp., and Achromobacter xylosoxidans could decolorize Congo red (25 mg/L) 93%, 62%, and 94%, respectively [102]. Bacterium Alishewanella spp. was reported for azo dyes degradation [103]. Mani et al. [104] reported Shewanella oneidensis for decolorization of acid orange 7 [Figure 3]. Lysinibacillus fusiformis was reported for degrading methyl red [105]. Saha and Rao [106] studied the degradation of Reactive Orange 16 up to 89% by Bacillus flexus. In a report, halo-alkaliphilic bacterium Nesterenkonia lacusekhoensis was reported for decolorization of Reactive Red-35 dye [107]. Akansha et al. [108] reported Bacillus stratosphericus for biodegradation of reactive orange 16 [Figure 4].
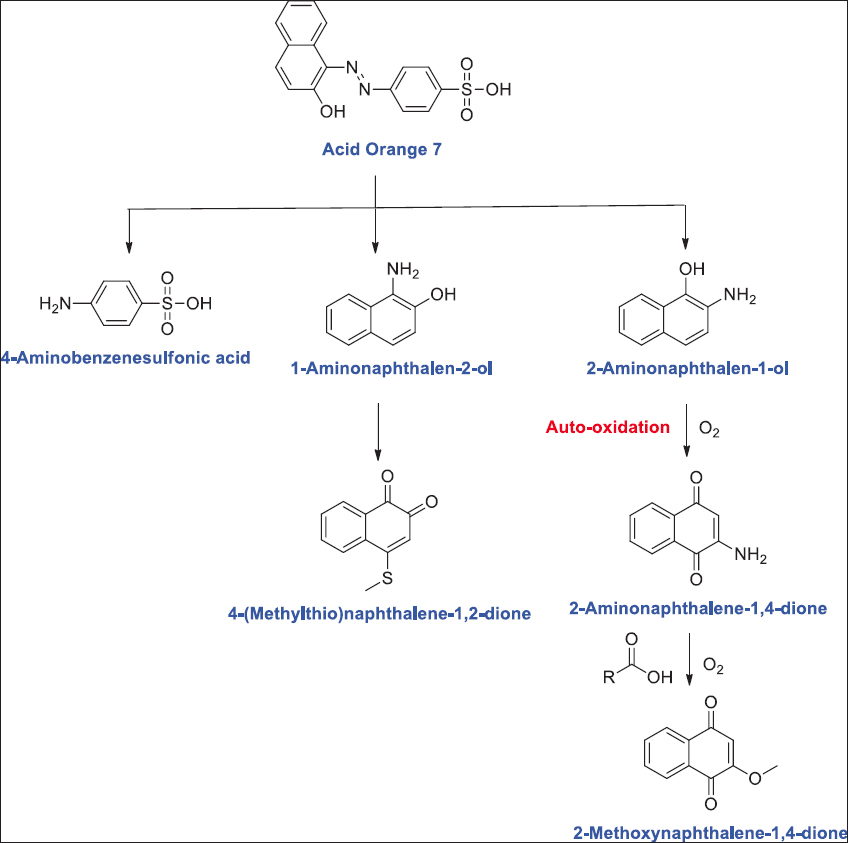 | Figure 3: Acid Orange 7 biodegradation pathways by Shewanella oneidensis. Source: Adapted with permission from Mani et al. [104]. [Click here to view] |
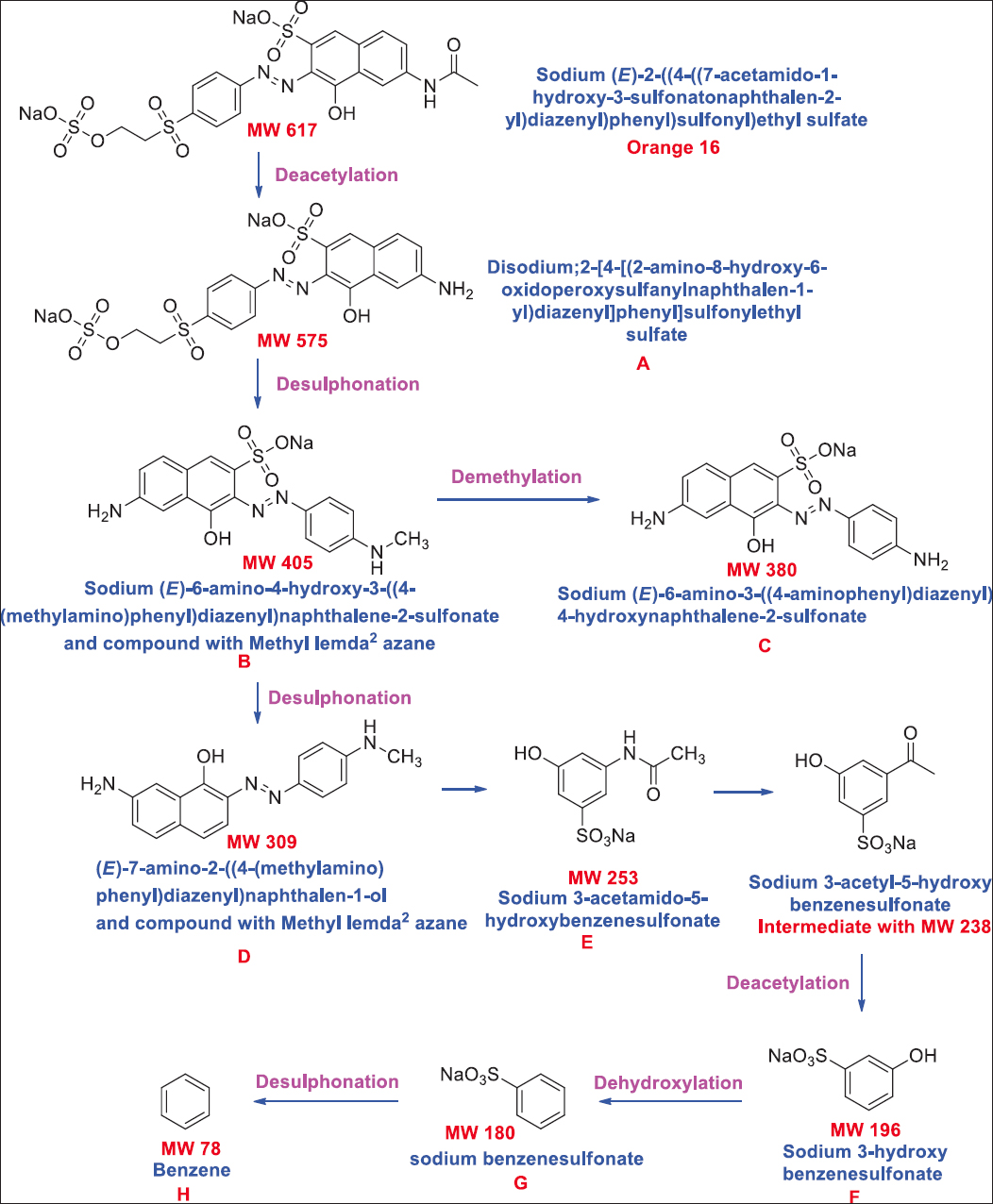 | Figure 4: Biodegradation pathway of Reactive Orange 16 through Bacillus stratosphericus. Source: Adapted with permission from Akansha et al. [108]. [Click here to view] |
4.3.3. Yeast
Yeast can grow fast and withstand adverse environmental conditions [109]. Trichosporon beigelu was capable to decolorize Navy blue HER, Golden Yellow 4BD, Red HE7B, Green HE 4BD, Orange HE2R, and Malachite green [22] whereas Candida krusei was able to decolorize Reactive Brilliant Red K-2BP, Acid Mordant Yellow Reactive, Weak Acid Brilliant Red B, Reactive Black KN-B, Reactive Brilliant Blue X-BR, Acid Mordant Light Blue B Reactive, Acid Mordant Red S-80, and Reactive Turquoise Blue KN-G [109]. Yeast C. krusei, isolated from textile wastewater, was reported for degrading Basic Violet 3 by 74% [110]. About 90% Acid Red B decolorization under aerobic conditions was reported by Pichia spp. [111]. In a report, Acid Brilliant Scarlet GR was detoxified by Candida tropicalis [112]. Tan et al. [113] reported Magnusiomyces ingens for degrading Acid Red B. In a report, Acid Red B was degraded by C. tropicalis [114]. Tan et al. [115] reported salt tolerant Scheffersomyces spartinae for the detoxification of Acid Scarlet 3R under anaerobic conditions. A recent study had reported complete decolorization of Reactive Orange 16 by Pichia kudriarzenii [116]. Cyberlindnera samutprakarnensis, the salt tolerant yeast, was reported for decolorization of Acid Red B by 97% within 18 h of incubation [117]. In a report, Galactomyces geotrichum was reported for degrading Acid Scarlet GR under aerobic conditions [118]. Similarly, halotolerant, Sterigmatomyces halophilus was reported for complete detoxification of Reactive Black 5 within 24 h of incubation [119]. In another report, C. tropicalis was reported for the Acid Red B degradation [120].
4.3.4. Algae
Some microbial groups have not been studied extensively for their degradation abilities concerning pollutants such as synthetic dyes and xenobiotics. Cyanobacteria (blue-green algae) have distributed ubiquitously, but there is scant information about their ecosystem functioning role, including recalcitrant compounds degradation such as dyestuffs and dye [121]. Chroococcus minutus, Gloeocapsa pleurocapsoides, and Phormidium ceylanicum are some of the algae reported on dye decolorization [122]. In a report, detoxification of monoazo and diazo dye was reported by the Nitzschia perminuta and Scenedesmus bijugatus [123]. Khataee et al. [124] reported Chara spp., the macroalgae for the detoxification of Malachite Green solution. In a report, from a thermal spring environment, Methylene Blue and Malachite Green were removed by Chlorella spp. and Chlamydomonas spp. [125]. In another report, Orange G Dye was detoxified by the microalgae Acutodesmus obliquues [126]. Chia et al. [127] reported Scenedesmus quadricauda for the degradation of Indigo Blue dye. Congo red dye detoxification was reported by green algae Chlorella spp., Chlorella vulgaris, Haematococcus spp., Scenedesmus officinalis, Scenedesmus obliquuss, and S. quadricauda [128]. In similar report, Methylene Blue and Malachite Green were decolorized by the Desmodesmus spp. [129]. C. vulgaris was reported for Indigo Blue dye [130].
4.3.5. Microbial consortium
In various studies, microbial consortia have shown greater effectiveness as compared to pure strains [21]. The mixed microbial population has shown greater efficiency of synthetic dyes decolorization than pure culture because of synergistic metabolic activities of microbial communities [131]. Each strain in the consortium may target different sites in the dye molecules or may utilize degraded metabolites generated by one strain followed by degradation by another strain [132]. Thus, using mixed microbial cultures dyes, biodegradation could be improved due to the synergistic effect [133]. Consortium development which can survive in the effluent by utilizing the components as a source of carbon, nitrogen, and energy would render the whole technique economically friendly [21]. Different microbial consortiums were reported for the remediation of various dyes [Table 1]. Tony et al. [134] showed 50–60% of biodegradation of dyes including Congo red, Bordeaux, Blue BCC, and Ranocid Fast Blue by bacterial consortium of Bacillus cereus, B. megaterium, B. pumilus, B. vallismortis, and B. subtilis. In a report, bacterial consortium of Bacillus odysseyi, Morganella morganii, and Proteus spp. was reported for textile dyes decolorization [135].
Table 1: Dye remediation using microbial consortium.
| Consortium | Dye | References |
|---|---|---|
| Bacillus vallismortis, B. cereus, B. pumilus, B. subtilis, and B. megaterium | Blue BCC, Ranocid fast blue, Bordeaux, Congo red | Patil et al. [135] |
| Brevibacillus laterosporus and Galactomyces geotrichum | Golden yellow HER | Waghmode et al. [136] |
| Klebsiella, Buttiauxella, and Bacillus | Methyl red | Cui et al. [137] |
| Citrobacter freundii, Moraxella osloensis, and Pseudomonas aeruginosa | Mordant black 17 | Karunya et al. [138] |
| Provedencia rettgeri, and Pseudomonas spp. | Reactive Orange 16 | Lade et al. [139] |
| Escherichia coli, Salmonella spp., Staphylococcus aureus, Proteus spp., Pseudomonas spp., and B. subtilis | Congo red | Holey [140] |
| Bacillus spp., B. subtilis, B. cereus, B. mycoides, Micrococcus spp., and Pseudomonas spp. | Green dye, red dye, black dye, and yellow dye | Mahmood et al. [141] |
| Pseudomonas stutzeri and Acinetobacter baumannii | Congo red and gentian violet | Kuppusamy et al. [142] |
| Aeromonas spp., Bacillus spp., Neisseria spp., and Vibrio spp. | Novacron brilliant blue fn-r, bezema yellow s8-g, novacron super black G | Karim et al. [143] |
| Barnettozyma californica, Sterigmatomyces halophilus, and Yarrowia spp. | Remazol brilliant blue R | Ali et al. [144] |
| Zobellella, Rheinheimera, and Marinobacterium | Direct blue B, acid violet 7, acid black ATT | Guo et al. [145] |
| Scenedesmus obliquus, and Oscillatoria spp. | Reactive orange 122, reactive red 194 | El-Sheekh et al. [146] |
| Bacillus odyssey, Morganella morganii and Proteus spp. | Red HE3B | Phugare et al. [147] |
| Providencia spp. and Pseudomonas aeuroginosa | Red HE3B | Phugare et al. [147] |
| Anoxybacillus spp., Clostridium spp., and Bacillus spp. | Direct Black G | Chen et al. [148] |
| B. subtilis, B. subtilis, and B. cereus | Mixture of azo dyes | Thiruppathi et al. [149] |
| Bacillus flexus, Proteus mirabilis, and Pseudomonas aeruginosa | Indanthrene blue RS | Kumar and Mohanty [150] |
| Lysinibacillus spp., Bacillus spp., Bacillus spp., Bacillus spp., Bacillus spp., and Ochrobacterium spp. | Reactive violet 5R | Jain et al. [151] |
| Galactomyces geotrichum and Bacillus spp. | Brilliant blue G | Jadhav et al. [152] |
| B. cereus, Pseudomonas putida, Pseudomonas fluorescence, and Stenotrophomonas acidaminiphila | Acid red 88 | Khehra et al. [153] |
| Aeromonas caviae, Proteus mirabilis, and Rhodococcus globerulus | Acid Orange 7 | Joshi et al. [154] |
| Enterobacter dissolvens and Pseudomonas aeruginosa | Acid maroon V | Patel et al. [155] |
| Stenotrophomonas rhizophila, Sphingomnas echinoides, Pseudarthrobacter oxydans, and Gordonia westfalica | Reactive black-5 | Eskandari et al. [156] |
| Pseudomonas aeroginosa, Stenotrophomonas maltophila, and Proteus mirabilis | Direct black 22 | Mohana et al. [157] |
| Aspergillus ochraceus and Pseudomonas spp. | Rubine GFL | Lade et al. [158] |
| Pseudomonas aeruginosa and Bacillus circulans | Reactive Black 5 | Dafale et al. [159] |
| Pseudomonas aeruginosa, Rhodobacter sphaeroides, Proteus mirabilis, and Bacillus circulance | Remazol black-B | Dafale et al. [160] |
| Enterococcus faecalis and Klebsiella variicola | Reactive red 198 | Eslami et al. [161] |
| Alcaligenes faecalis, Sphingomonas spp., B. subtilis, Bacillus thuringiensis, and Enterobacter cancerogenus | Direct blue-15 | Kumar et al. [162] |
| Anoxybacillus pushchinoensis, A. kamchatkensis, and Anoxybacillus flavithermus | Reactive black 5 | Deive et al. [163] |
| Dichotomomyces cejpii and Phoma tropica | Congo red, methyl red, reactive blue | Krishnamoorthy et al. [164] |
| Bacillus spp., Stenotrophomonas spp., Pseudomonas spp., and Alcaligenes spp. | Procion red H-3B | Shah and Bera [165] |
| Pseudomonas aeruginosa, Enterobacter spp., and Serratia marcescens | Reactive red 120 | Manogaran et al. [166] |
| Penicillium spp. and Sphingomonas xenophaga | Reactive brilliant red X-3B | Gou et al. [167] |
B. cereus: Bacillus cereus, B. pumilus: Bacillus pumilus, B. subtilis: Bacillus subtilis, B. megaterium: Bacillus megaterium.
The consortium of Brevibacillus laterosporus and G. geotrichum was reported for Golden Yellow HER decolorization [136]. In another report consortium, bacterial strains Klebsiella, Buttiauxella, and Bacillus were reported for the removal of Methyl Red under aerobic conditions [137]. Karunya et al. [138] developed a microbial consortium of Citrobacter freundii, Moraxella osloensis, Pseudomonas aeruginosa, and P. aeruginosa which helps in the Mordant Black 17 decolorization. Lade et al. [139] used microbial consortium consisting of Provedencia rettgeri and Pseudomonas spp. for decolorization of Reactive Orange 16. The consortium could decolorize 1110 mg/L of Reactive Orange 16 by about 99%. Another study by Holey [140] reported a where consortium which comprised E. coli, Salmonella spp., Staphylococcus aureus, Proteus spp., Pseudomonas spp., and B. subtilis and had the ability to decolorize Congo Red (10 mg/L) up to 98% within 96 h of incubation. In another study, the results indicated that the consortium ability to decolorize the green, red, yellow, and black dyes was higher as compared to single strains application (Bacillus spp., B. subtilis, B. cereus, B. mycoides, Micrococcus spp., and Pseudomonas spp.). The consortium was capable to decolorize green (84%), red (84%), yellow (85%), black (85%), and mixed dyes (82%) within 24 h while individual strain required 72 h [141].
In an investigation, bacterial consortium of novel and indigenous strains, namely, Pseudomonas stutzeri and A. baumannii was reported for textile dyes decolorization [142]. In case of monoculture (Aeromonas spp., Bacillus spp. Neisseria spp., and Vibrio spp.), percentage of decolorization varies from no visible decolorization to highest 90% decolorization (Novacron Brilliant Blue FN-R) whereas the percentage decolorization of bacterial consortium varies from 65% (Benzema Yellow S8-G) to 90% (Novacron Super Black G and Novacron Brilliant Blue FN-R) [143]. Reactive azo dyes were also reported to be degraded by the consortium of yeast, that is, Barnettozyma californica, S. halophilus, and Yarrowia spp. [144]. Guo et al. [145] showed the detoxification of Acid Black ATT, Direct Blue B, and Acid Violet 7 by the consortium containing halotolerant microbes, namely, Zobellella, Rheinheimera, and Marinobacterium. Consortium of cyanobacteria and green algae, that is, S. obliquus and Oscillatoria spp. was reported for degradation of azo dyes including Reactive Orange 122 and Reactive Red 194 [146].
5. MECHANISM OF DYE BIODEGRADATION
The plant and microbial systems possess efficient enzymatic systems which could be used for bioremediation. Therefore, it is important to find the mechanism of biotransformation followed by the organism and probable enzymes which are involved in biochemical complex reactions. The enzymatic treatment methods positively affect the environment as they pose a low chance of biological contamination. Enzymes of both bacterial as well as fungal origin such as lignin peroxidase, laccase, and manganese peroxidase have the capacity to metabolize xenobiotic compounds [168]. Peroxidase enzyme purified from plant species which includes Saccharum spontaneum and Ipomoea palmata is used for efficient decolorization of various textile dyes [169]. Enzymes like horseradish peroxidase have been immobilized and utilized for the treatment of effluents of textile mills and paper industries [170].
Laccase belongs to the multicopper oxidases group which has low substrate specificity and is highly capable of degrading the spectrum of xenobiotic compounds and aromatic as well as non-aromatic substrates [171]. These catalysts have good bioremediation potential at the same time. It does not require available oxygen as an electron acceptor, which makes them highly applicable in many biotechnological processes [172]. Enzymes can degrade phenolic compounds and aromatic azo compounds. Cu2+ is used as mediator to oxidize aromatic amines. First prokaryotic laccase has been reported by Azospirillum lipoferum [173]. Laccase basically catalyzes azo dyes decolorization by non-specific free radical without the mutagenetic and toxic aromatic amines formation. Pseudomonas syringae and Pedomicrobium spp. have shown laccase like activity [174]. Reports are available for purified laccase enzyme extracted from Bacillus spp. and Pseudomonas desmolyticum that can decolorize several textile dyes efficiently [175,176]. Fungal strain Podoscypha eleganscan decolorize five azo dyes (Congo Red, Direct Blue 15, Orange G, Rose Bengal, and Direct Yellow) efficiently [171]. Similarly, fungal strains of Aspergillus oryzae, Trametes versicolor, and Paraconiothyrium variable can decolorize azo dyes through the laccase enzyme production [177].
Azoreductase also known as azobenzene reductase is a reducing enzyme. These catalysts can degrade azo dye into colorless amines through the process of reductive cleavage. The whole process requires NADH or FADH [178] which acts as an electron donor in a redox reaction [45]. Bio-treatment of azo dye containing effluent and microbial azoreductase has been observed to play a major role. Sometimes, under unfavorable environmental conditions, few usual cellular enzymes might get converted into dye degrading enzyme example flavin reductase produced by E. coli acts as azoreductase [179]. Microbes such as B. subtilis, Pseudomonas spp., and S. aureus have been found to decolorize azo dyes (Methyl Red, Disperse Blue, and Acid yellow) through the production of azoreductase [180-182]. Agrawal et al. [183] reported Providencia spp. for degrading Acid Black 210 through the production of azoreductase [Figure 5].
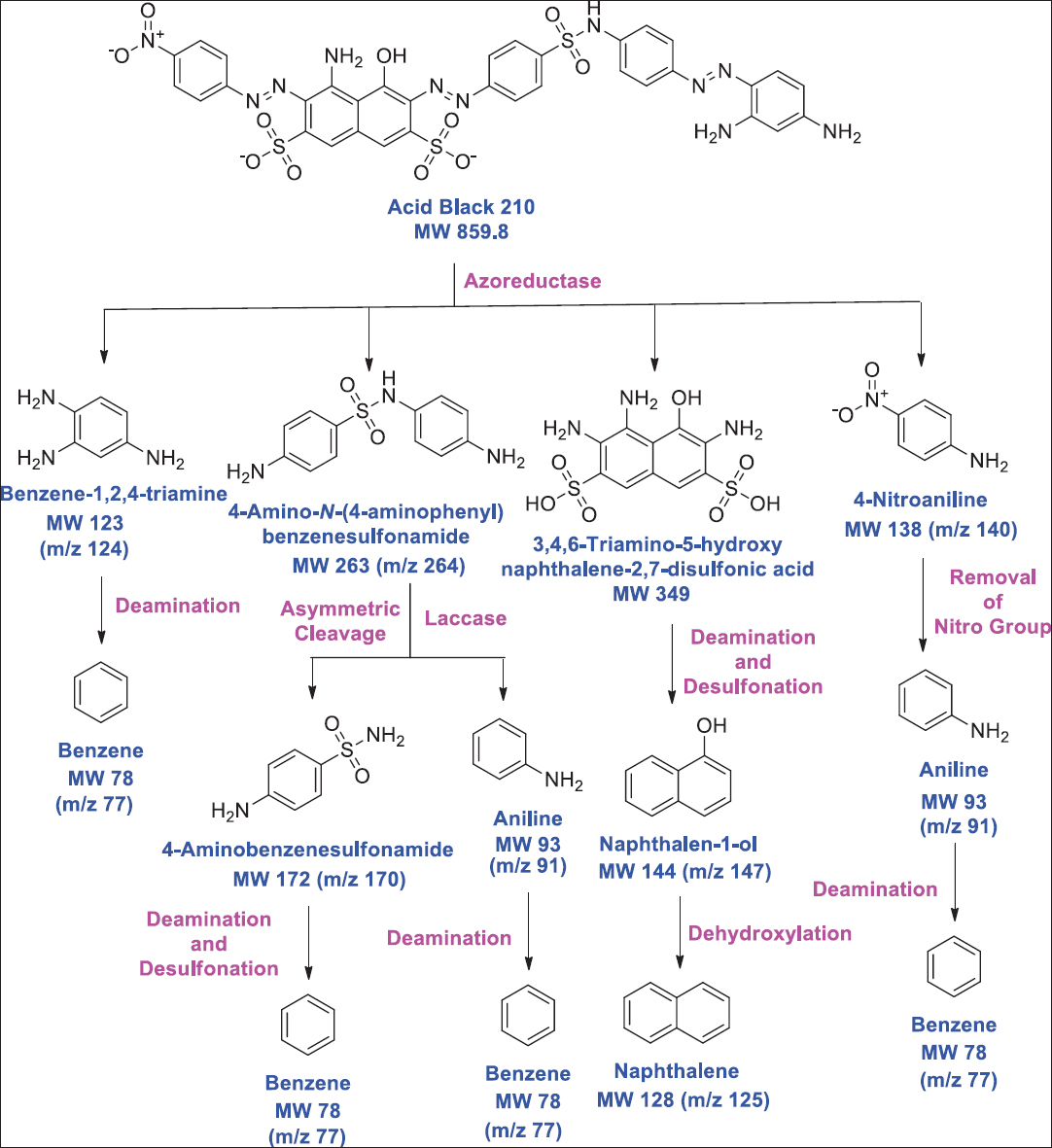 | Figure 5: Azoreductase-mediated biodegradation of azo dye acid black 210 through Providencia sp. Source: Adapted with permission from Agrawal et al. [183]. [Click here to view] |
Peroxidase is related to the group of oxidoreductases, especially which act on peroxide acting as the electron acceptor. Knowledge of such factors will influence the degradation activity which will facilitate the bioreactor development for bioremediation of industrial waste. The fungus’s efficiency to degrade azo dyes is related to the exo-enzymes formation such as peroxidases and phenol oxidases. Peroxidases can catalyze the breakdown of hydrogen peroxide into water and molecular oxygen [184]. These enzymes have a heme group attached to the active site [185]. Both manganese peroxidases and lignin have the same reaction mechanism (catalytic enzyme causes oxidation of H2O2 to an oxidized state). Basically, lignin peroxidases help in the oxidation of non-phenolic aromatic compounds whereas manganese peroxidases oxidize Mn2+ to Mn3+ and this Mn3+ is responsible for the oxidation of numerous phenolic compounds [186]. The first peroxidase was obtained from Phanerochaete chrysosporium [187]. Various microorganisms involved in dye decolorization with peroxidases activity include Rhodococcus jostii [188], I. lacteus [189], Thermomonospora curvata [190], B. subtilis [191], Enterobacter ignolyticus [192].
6. FACTORS AFFECTING BIOREMEDIATION
The whole ecosystem consists of a dynamic environment with various abiotic factors such as temperature, metals, salts, pH, and the presence of oxygen. Microbes play a key role in the carbon, nitrogen, and sulfur cycle and are greatly influenced by any change in these factors thus, affecting the decomposition process [Table 2]. Thus, it is very important to analyze the effect of these parameters on xenobiotics degradation. Knowledge of these factors which play a pivotal role in degradation activity will likely facilitate the development of bioreactors for the bioremediation of industrial waste.
Table 2: Factors affecting the microbes mediated remediation of dyes.
| Microbes | Dye | pH | Temperature (°C) | Initial conc. of dye (mg/L) | Static/Agitation | References |
|---|---|---|---|---|---|---|
| Acinetobacter baumannii | Reactive red | 7.0 | 37 | 500 | Agitation | Unnikrishnanet al. [226] |
| Acinetobacter spp. | Reactive orange 16 | 7.0 | 40 | 500 | Static | Meerbergenet al. [218] |
| Aeromonas hydrophila | Reactive Black 5 | 7.0 | 35 | 100 | Static | El Bouraie and El Din [216] |
| Aeromonas veronii | Methyl orange | 7.0 | 32 | 1000 | Static | Mnifet al. [227] |
| Alcaligenes aquatilis | Synazol red 6HBN | 7.0 | 37 | 10 | Static | Ajazet al. [219] |
| Alcaligenes faecalis | Novacron super black G | 8.0 | 37 | 200 | Static | Hossenet al. [203] |
| Anoxybacillus spp. | Direct black G | 7.2 | 55 | 400 | Static | Chenet al. [228] |
| Aspergillus flavus | Reactive red 198 | 4.0 | 3 | 50 | Agitation | Esmaeili and Kalantari [196] |
| Aspergillus niger | Crystal violet | 5.5 | 30 | - | Static | Aliet al.[200] |
| Bacillus algicola | Yellow azo dye | 8.0 | 25 | - | - | Chukowryet al. [217] |
| Bacillus cereus | Novacron super black G | 8.0 | 37 | 200 | Static | Hossenet al. [203] |
| Bacillus fusiformis | Acid orange 10 | 9.0 | 37 | 150 | Static | Kolekaret al. [229] |
| Bacillus spp. | Reactive red 239 | 10.0 | 30 | 250 | Agitation | Guadieet al. [201] |
| Bacillus subtilis | Reactive blue 160 | 7.0 | 35 | 500 | Agitation | Barathiet al. [220] |
| Bacillus vallismortis | Aniline blue | 6.0 | 70 | - | - | Zhanget al. [212] |
| Bacillus vietnamensis | Malachite green | - | 37 | 50 | Static | Kabeeret al. [230] |
| Bjerkandera adusta | Lanaset grey G | 6.0 | 40 | 150 | Agitation | Daâssiet al. [231] |
| Comamonas spp. | Direct red 5B | 6.5 | 40 | 1100 | - | Jadhavet al. [232] |
| Coriolopsis gallica | Lanaset grey G | 6.0 | 45 | 150 | Agitation | Daâssiet al. [231] |
| Enterobacter hormaechei | Reactive yellow 145 | 7.0 | 37 | 100 | Agitation | Thangarajet al. [221] |
| Enterobacter hormaechei | Reactive yellow 145 | 7.0 | 37 | 100 | Agitation | Thangarajet al. [221] |
| Enterobacter spp. | Reactive black 5 | 7.0 | 37 | 200 | Static | Wanget al. [195] |
| Exiguobacterium spp. | Navy blue HE2R | 7.0 | 30 | 50 | Static | Dhanveet al. [233] |
| Geotrichum candidum | Reactive blue 5 | - | 30 | 120 | Agitation | Kimet al. [234] |
| Halomonas glaciei | Reactive red 2 | 8.1 | 30 | 1000 | Static | Balamuruganet al. [235] |
| Halomonas variabilis | Reactive red 2 | 8.1 | 30 | 1000 | Static | Balamuruganet al. [235] |
| Issatchenkia orientalis | Direct black 22 | 7.0 | 32 | 500 | Agitation | Jafariet al. [236] |
| Kocuria rosea | Methyl orange | 6.8 | 30 | 50 | Static | Parshettiet al. [94] |
| Lactobacillus paracase | Acid black | 6.0 | 30 | 100 | Agitation | Huanget al. [199] |
| Lysinibacillus fusiformis | Methyl red | - | 30 | 100 | Agitation | Sari and Simarani [105] |
| Lysinibacillus sphaericus | Drimaren red CL-5B | 8.0 | 37 | 100 | Agitation | Srinivasan and Sadasivam [237] |
| Lysinibacillus spp. | C.I. Remazol red | 7.0 | 30 | 250 | Static | Sarataleet al. [197] |
| Micrococcus luteus | Direct orange 16 | 8.0 | 37 | 100 | Static | Singhet al. [215] |
| Micrococcus yunnanensis | Methyl orange | 7.0 | 30 | 100 | Agitation | Carolinet al. [205] |
| Moraxella osloensis | Mordant black 17 | 7.0 | 35 | 100 | Static | Karunyaet al.[214] |
| Nesterenkonia lacusekhoensis | Methyl red | 11.5 | 30 | 50 | Static | Bhattacharyaet al. [238] |
| Nesterenkonia lacusekhoensis | Reactive violet 1 | 11.5 | 27 | 200 | Agitation | Prabhakaret al. [239] |
| Ochrobacterium spp. | Reactive violet 5R | 7.0 | 37 | 200 | Static | Jainet al. [151] |
| Ochrobactrum anthropic | Reactive black 5 | 7.0 | 30 | 400 | Static | Chenget al. [240] |
| Penicillium ochrochloron | Cotton blue | 6.5 | 25 | 50 | Static | Shedbalkaret al. [209] |
| Pichia kudriavzevii | Acid red B | 5.0 | 33 | 100 | Agitation | Fenget al. [241] |
| Pseudomonas aeruginosa | Direct orange 39 | 7.0 | 60 | 50 | Agitation | Jadhavet al. [225] |
| Pseudomonas azoreducens | Reactive green | 7.0 | 30 | 500 | Static | Meerbergenet al. [218] |
| Pseudomonas putida | Orange II | 8.0 | 30 | 100 | Static | Kumaret al. [242] |
| Pseudomonas spp. | Reactive red 195 | 8.0 | 40 | 100 | Static | Khanet al. [198] |
| Sphingomonas paucimobilis | Methyl red | 9.0 | 30 | 750 | Agitation | Ayedet al. [211] |
| Thiosphaera pantotropha | Reactive yellow 145 | 7.0 | - | 100 | Static | Garget al. [243] |
| Trametes trogii | Lanaset gray G | 5.0 | 45 | 150 | Agitation | Daâssiet al. [231] |
| Trametes versicolor | Lanaset gray G | 5.0 | 45 | 150 | Agitation | Daâssiet al. [231] |
| Trichoderma harzianum | Cresol red | - | 25 | - | Agitation | Noret al. [244] |
| Trichosporon beigelii | Navy blue | 7.0 | 37 | 50 | Static | Sarataleet al. [22] |
| Bacillus stratosphericus | Reactive orange 16 | 7.0 | 35 | 150 | Static | Akanshaet al.[108] |
6.1. pH
In general, effective decolorization or degradation of dyes using bacteria takes place at basic or neutral pH, while yeast and fungi at neutral or acidic pH [67]. At pH below 4, H+ ions compete effectively with cations of dye, causing a reduction in efficiency of color removal, while at pH higher above this point charge, the biomass surface gets negatively charged, which attracts the dye positively charged cations through electrostatic force of attraction [193]. The study on 27 different dyes by 21 various basidiomycetes reveals that optimum pH was found to be in the 3–5 range for dye decolorization [194]. A study demonstrated Enterobacter spp. decolorizes Reactive Black 5 at pH 7.0 [195]. The study revealed that decolorization and degradations of Scarlet R by Proteus vulgarius and Micrococcus glutamicus occurred in the range of pH 7–8 with optimum pH 7 [90]. In a study, bacterial mixtures consisting of total of six bacterial species, namely, Bacillus spp. (four strain), Lysinibacillus spp., and Ochrobacterium spp. were reported for decolorization and removal of azo dye-Reactive Violet 5R at neutral pH i.e, 7.0 [151]. In another investigation, the bacterium A. flavus sorted out from the effluent disposal area soil was reported for degrading Reactive Red 198 effectively at low pH [196].
Another report has reported bacterium Lysinibacillus spp. isolated from the textile industrial area which was degrading and decolorizing the toxic sulfonated azo dye C.I. Remazol Red at pH 7.0 [197]. In a study by Khan et al. [198], Reactive Red 195 dye was degraded by the bacterial consortium consisting of, Bacillus spp., Pseudomonas spp., and Ochrobactrum spp. at pH 8.0. In an investigation, the bacterium Lactobacillus paracase isolated form the deep sea sediments were reported for removal of Acid Black azo dye between the pH range from 5.0 to 7.0 [199]. In another study, the most effective pH for the Crystal Violet decolorization by the fungi A. niger was reported 5.5 [200]. Bacillus spp., isolated from an alkaline lake, was reported for degradation of Reactive Red 239 dye. The most efficient pH for dye degradation by the bacterium was 10 [201]. In another investigation, Enterobacter spp. was degrading Crystal Violet at pH 6.5 [202]. In a report, bacteria obtained from textile industry released effluent were identified as Alcaligenes faecalis, Bacillus spp., and B. cereus which were reported for biodegradation of Novacron Super Black G dye at pH 8.0 under static conditions [203]. Javadzadeh and Asoodeh [204] isolated Bacillus spp. from the gut of termite which was having indigo dye biodegrading properties. The bacterium was reported for biodegrading the dye at pH 8.0. In an investigation, the microbial species Micrococcus yunnanensis was reported for degrading Methyl Orange at pH 7 [205].
6.2. Temperature
Temperature is a vital factor and plays an imperative role in the environment during the process of biodegradation. The metabolic activity of microorganisms is greatly affected by temperature. It is commonly experiential that the decomposition process is faster in summer as compared to winter as a warmer climate favors the growth and multiplication of various microorganisms [206]. However, the process is not the same after a certain temperature. Beyond this optimum temperature, there is a reduction in growth, metabolic activity, and deactivation of enzymes which ultimately lead to a decline in the decolorization process [193]. Thus, various studies conclude that biodegradation of dye by the microorganism is possible at ambient temperature which is responsible for their metabolic activities and reproduction [207]. Various microorganisms require different ambient temperatures for growth, with most growing at a temperature range of 25–35°C [208]. In a study by Shedbalkar et al. [209], the ambient temperature required to decolorize Cotton Blue by Penicillium ochrochloron was found to be 25°C. Enterobacter spp. could decolorize Reactive Black 5 at an optimum temperature of 37°C and on further increasing the temperature to 42°C there was a drastic decrease in decolorization activity [195]. In a study, Pseudomonas spp. was reported for degrading Congo red dye along with textile industry released effluent at 40°C temperature [210]. An isolate Sphingomonas paucimobilis was reported for bioremoval of Methyl Red dye between a broad range of temperatures i.e. 4–40°C [211]. In another report, B. vallismortis isolated from the disposal site of the textile industry was reported for degrading triphenylmethane dyes, including Aniline blue, Malachite Green, and Brilliant Green at high temperature range of 70°C [212].
A study has reported, Reactive Black-5 dye was reported to be degraded by the halotolerant bacterial strain Pseudomonas spp. at a temperature 25°C [213]. Similarly, the bacterium M. osloensis was reportedly degrading Mordant Black 17 at 35°C temperature [214]. In a report, Direct Orange 16 was degraded at 37°C temperature by bacterial isolate Micrococcus luteus [215]. Reactive Black 5 dye was reported to be biodegraded by the Aeromonas hydrophila ate 35°C temperature [216]. In a study, the bacterial strain Bacillus algicola was reported for decolorizing red, yellow, and blue dye at temperatures 25°C, 35°C, and 45°C. The highest degradation was observed at 25°C temperature [217]. In a similar report, two bacterial strains, namely, Acinetobacter and Klebsiella were decolorizing the azo dyes, that is, diazo dye Reactive Green 19 and monoazo dye Reactive Orange 16 at a temperature range of 20–40°C [218]. Ajaz et al. [219] reported Alcaligenes aquatilis for decolorization of Synazol Red 6HBN dye at 37°C temperature in 4 days [Figure 6]. In a report, B. subtilis was reported for decolorizing Reactive Blue 160 dye at 35°C temperatures [220]. Enterobacter hormaechei isolated from textile effluent was reported to degrade Reactive Yellow 145 and Reactive Red 180 at a temperature 37°C [221].
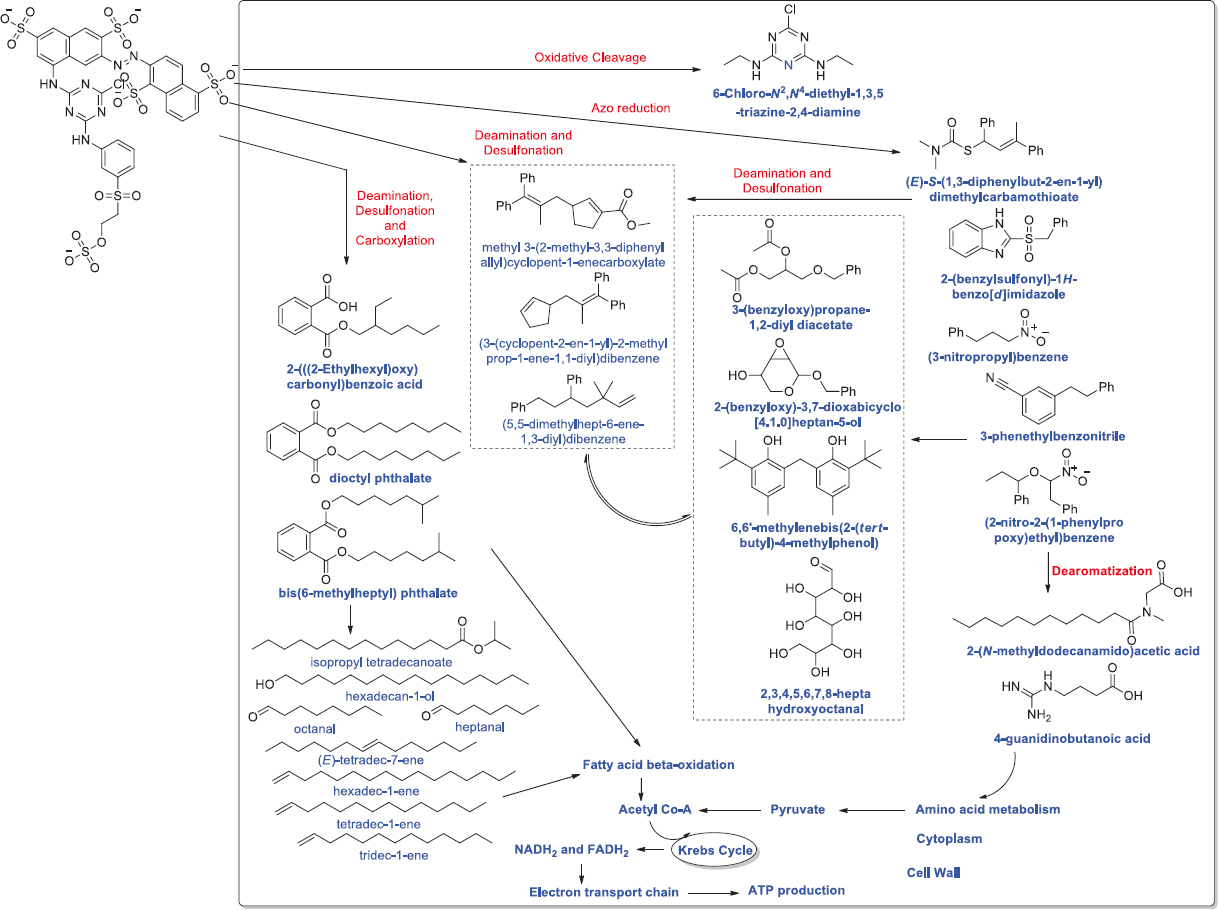 | Figure 6: Biodegradation of Synazol Red HF 6BN dye Alcaligenes aquatilis. Synazol Red HF 6BN enters the cell of A. aquatilis through unknown process. After the entrance dye is processed enzymatically into several end products. First, the azo group of the dye is reduced which is followed by cleavage reaction to form various end products. Second, the desulfonation and oxidative deamination results in synthesis of pyrrolo[1,2-a] pyrazine-1,4-dione derivative which can be used as substrates in amino acid metabolism. The amino acid catabolism can synthesize pyruvate (3C compound) which can be converted into acetyl-CoA. The acetyl-CoA undergoes Krebs cycle to produce NADH2 and FADH2 (substrates of electron transport chain). Moreover, dye desulfonation, oxidative deamination, and carboxylation lead to produce phthalate derivatives, which can be transformed into different fatty acids and aldehydes. The phthalate, fatty acids, and aldehydes can directly/indirectly enter into fatty acid oxidation reactions (β-oxidation) to produce acetyl-CoA, NADH2 and FADH2. Source: Adapted with permission from Ajaz et al. [219]. [Click here to view] |
6.3. Initial Dye Concentration
The impact on microbial insisted decolorization of the dye initial concentration was investigated. Studies show that there is a lowering in the efficiency of dye decolorization by microbes with an initial concentration increase of dye. This is because the dye toxicity increases at a higher concentration which inversely affects the growth of the microbial cells [193]. A similar report was demonstrated by Parshetti et al. [222] which indicated that higher concentration Malachite Green (100 mg/L) was toxic to K. rosea. Jirasripongpun et al. [223] found that Reactive Red 195 at concentrations 50 and 100 mg/L had a lethal effect on Enterobacter spp. and was not able to grow. A report on Congo red decolorization by Bacillus spp. reported that the decolorization rate declined with the dye concentration increase [224]. In a report, P. aeruginosa obtained from the dyestuff contaminated sediments was reported for 93.06% decolorization of Direct Orange 39 with 50 mg/L concentration within 45 ± 5 min and the maximum concentration degraded by the strain was 1.5 g L−1 with 60% decolorization [225]. The consortium of Providencia spp. and Pseudomonas aeuroginosa was reported for 100% bioremediation of dye Red HE3B at 50 mg/L initial concentration and as the dye concentration rises to 200 mg/L and 250 mg/L the decolorization decreases to 85% and 70%, respectively [147].
A study reported Pseudomonas putida was decolorizing dye Orange II at maximum up to 1000 mg/L initial concentration and maximum decolorization was achieved at 100 mg/L concentration [242]. Shah et al. [53] reported that as the concentration of methyl orange was increased (3, 4, and 5 g/L), the incubation time required for decolorization was varied from 66 to 90 h. In another report, Pichia kudriavzevii was reported for degrading Acid Red B dye with maximum initial concentration 400 mg/L within 40 h and maximum decolorization was achieved with 100 mg/L initial concentration within 40 h [241]. Saroj et al. [245] concluded, the fungal consortium containing Penicillium oxalicum, A. niger, and A. flavus strains was able to degrade three different azo dyes, namely, Direct Blue 15, Direct Red 75, and Acid Red 183, with the initial concentration range 200–400 mg/L. All these three dyes were degraded at lower initial concentrations by the fungal consortium. Bacterium Aeromonas veronii sorted out from acclimated textile effluent was reported to decolorize the azo dye up to 1000 mg/L initial concentration [227].
Reactive Blue 4, the anthraquinone dye, was degraded by the bacterial granules up to 1000 mg/L initial concentration [246]. In a report, thermophilic microflora was reported for detoxification of azo-dye Direct Black G with an initial concentration 600 mgL−1 [247]. In another report, Reactive Black-5 dye decolorization through bacterial (Gordonia, Pseudoarthrobacter, Sphingomonas, and Stenotrophomonas) consortium was tested with three different initial concentrations, that is, 25, 50, and 100 mg/L. The best dye decolorization by the consortium of bacterial strains was found at 50 mg/L initial concentration [156]. Amin et al. [248] reported Bacillus spp. to detoxify the diazo dye at an initial concentration of 100 mg/L. In a study, Ochrobactrum anthropic from textile wastewater was reported for biodegradation of Reactive Black 5 at the initial concentration of 400 mg/L [240].
6.4. Sodium Chloride Concentration
Effluents from the textile industry contain various salts or metal ions, acids, and alkalis as impurity in addition to dyes [67]. The salt concentration varies from 15 to 20% and has been calculated in dye industry wastewater. Thus, a microbial strain that can tolerate high concentrations of salt facilitates the degradation of dye wastewater. The biological treatment of the dye effluent containing various components, and identification of salt tolerant microorganisms is important. Rudakiya, Pawar [249] have shown the decolorization efficacy of a bacterial strain using salt concentrations up to 10%. In their study, the result come out to be that 6% salinity was effective in promoting both cell biomass and degradation of Reactive Orange 16. A similar study was done on Shewanella marisflavi and S. algae algae which were able to degrade single or mixed azo dyes at lower concentrations of NaCl (2–3%). It has been concluded that a lower level of salinity induces the activities of azoreductase, laccase, and NADH-DCIP reductase enzymes [250]. Cui et al. [98] reported that Klebsiella spp. was able to decolorize Methyl red, Orange I, Congo red, and Methyl orange efficiently over the salinity range (1–4%). In a study, M. luteus bacterium was reported for the detoxification of the Direct Orange 16 dye with 3% NaCl concentration in 6 h of incubation [215]. In another report, halotolerant S. marisflavi reported for decolorization of Xylidine Ponceau 2R under the 20% concentration of NaCl [251]. Song et al. [252] reported a yeast Pichia occidentalis for the Acid Red B dye biodegradation in 16 h with 30 g L−1 of NaCl. In another report, Bacillus spp. was biodegrading 96% of sulfonated dye, that is, methyl orange with 5–20 g/L of sodium chloride concentration [253]. Salt tolerant bacterium Halomonas was reported for the biodegradation of Toluidine Red dye where the concentration of NaCl was 5% [254]. Zhuang et al. [255] reported Methyl Orange and Reactive Yellow 84 decolorization by the Shewanella indica and Oceanimonas smirnovii. These strains were decolorizing the dye in the presence of NaCl (0–70 g L−1). Similarly, A. baumannii was reported for the 87% and 90% degradation of Reactive Black 5 and Reactive Blue 221, respectively, with 5% NaCl concentration [256].
6.5. Agitation/Static Condition
There are conflicting reports on azo dye decolorization through microbes under shaking/static conditions. According to studies, the decolorization rate increases by shaking culture conditions while other reports suggest static conditions. A higher rate of decolorization was observed during shaking due to the easy transport of oxygen and nutrients over static conditions [257,258]. On the other hand, decolorization by Pseudomonas spp. under agitation showed no decolorization while static culture conditions showed 96% Reactive Red 2 decolorization [88]. Similarly, Direct Red 81 and Reactive Red 120 decolorization by A. niger were more efficient under static conditions [259]. The rate of enzymatic activity was higher in the still condition [67]. Complete Navy Blue dye decolorization by Trichosporon beigelii was observed under static conditions whereas it declined to 30% under shaking conditions [22]. Another study concluded K. rosea for 100% Methyl Orange decolorization under the static conditions [94]. In a report, halophilic and halotolerant Halomonas variabilis and Halomonas glaciei were reported for degrading reactive textile dye in batch mode static condition [235]. Shah et al. [260] reported A. faecalis for degradation of Reactive Orange 13 under static anoxic condition. In a study, white rot fungi, that is, B. adusta, Coriolopsis gallica, Trametes trogii, and T. versicolor were reported for decolorizing Lanaset Gray G dye under the static conditions [231]. In agitation condition, Issatchenkia orientalis was reported for the five azo dyes decolorization, that is, Reactive Red 198, Reactive Orange 16, Direct Yellow 12, Direct Black 22, and Direct Blue 71 [236]. Nor et al. [244] reported Trichoderma harzianum for degrading Cresol Red dye under agitation conditions. In a study, Chaetomium globosum was tested for dye detoxification under stirred and static conditions and the report concluded that fungi were efficiently degraded the dye under agitation conditions in comparison with static conditions [261]. Bhattacharya et al. [238] reported under static conditions, halotolerant and alkalophilic N. lacusekhoensis was efficiently detoxifying Methyl Red. Similarly, A. baumannii from the sea sediments was reported for Reactive Red dye degradation at both agitation and static conditions [226]. In a comparative study, L. fusiformis was experimented for azoreductase degradation under both static and agitation conditions and results showed that under agitation conditions, dye degradation was better [105]. In a study, under static conditions, P. aeruginosa and Thiosphaera pantotropha were reported for decolorization and Reactive Yellow 145 dye detoxification [243].
6.6. Aerobic/Anaerobic Culture Conditions
Azo dyes are usually resistant to the attack of bacterial species under aerobic conditions [4]. Anaerobic degradation of dye has been more effective than aerobic, but the intermediates formed are carcinogenic and toxic in nature and must be degraded before being discharged to the main water stream [262]. Apart from few, all aromatic amines produced after azo dyes decolorization has recalcitrant properties under oxygen limiting condition [152]. However, the process when combined shows much more efficient output as reported by many authors [262]. They reported that the dye degradation by bacteria under anaerobic conditions is usually accompanied by the colorless aromatic amine production which is easily metabolized under aerobic conditions. The breakdown products are non-toxic in nature [139]. Bacterium P. aeruginosa was able to degrade, Navitan Fast Blue SSR, under an aerobic condition in the presence of glucose. The organism was also able to discolorate various other textile dyes [263]. Aerobic bacteria consisting of B. cereus, B. megaterium, B. pumilus, B. subtilis, and B. vallismortis were found to be efficient in decolorizing microbial strains individually as well as the mixture of dyes. Almost 80–90% decolorization was noticed in four out of six azo dyes (Congo red, Bordeaux, Blue BCC, and Ranocid fast blue) when present in mixture [134]. In a study, thermophilic microbial strains Anoxybacillus pushchinoensis, A. kamchatkensis, and A. flavithermus blend were reported for detoxifying 80% of Reactive Black 5 in aerobic conditions [163]. A comparative study has reported, microbial strain A. hydrophila was efficiently degrading the synthetic dye basic fuchsin, Crystal violet, Solophenyl red 3BL, Polar brilliant red B, Safranin, and Nigrosine under microaerophilic conditions in comparison to aerobic conditions [264]. Aerobic detoxification of Acid Scarlet GR was reported by a halotolerant yeast strain G. geotrichum [118]. A study has reported efficient degradation of dye Acid Red 14 by microbial strain Oerskovia paurometabola under anaerobic conditions [265]. In another report, Pseudomonas spp. and Clostridium spp. were reported to degrade the dye under aerobic conditions [266].
7. PHYTOTOXICITY AND MICROBIAL TOXICITY OF DYES AND THEIR BIODEGRADATION PRODUCTS
It is essential to determine whether dye degradation leads to dye detoxification. Microbial toxicity and phytotoxicity studies on dye and its biodegraded metabolites can confirm this. In phytotoxicity studies, experimental plant seeds are treated with the dye and its degraded metabolites. Effect on germination percentage, length of root, and shoot is measured, to compare with control (without dye and its biodegraded metabolites). Results are enumerated to determine whether the degraded metabolites are less toxic to growing seedlings than the dyes being studied. Likewise, the antimicrobial activity (toxicity) of the dye being studied can be compared with its degraded metabolites. Reactive Red 2 and its degraded metabolites were studied using Phaseolus mungo and Sorghum vulgare as model plants by Kalyani et al. [88]. The result observed concluded that the product formed after degradation was less toxic than Reactive Red 2. A similar result was observed when Malachite Green phytotoxicity and its degraded metabolites using P. mungo and Triticum aestivum as a model plant was performed [222]. Phytotoxicity study of Navy blue 2GL and its biodegraded product by Bacillus spp. on Sorghum bicolor and Triticum aestivium have shown similar results when compared with dye as control. There was 2–5% reduction in growth (root and shoot length) in the presence of degraded products than in distilled water [267]. The microbial toxicity study on Azobacter vinelandii showed a growth inhibitory zone of 1.2 cm against dye while no zone of inhibition was found around metabolites formed. The toxicity of crystal violet dye against E. coli was checked. The study reveals the toxic nature of crystal violet, but the product formed after degradation with Shewanella decolorationis NTOU1 was non-toxic to E. coli [268].
8. FUTURE PERSPECTIVES
The rampant industrialization has proliferated the huge amount of effluent containing mixture of dyes, metals and other hazardous materials. The degradation of dyes and other pollutants has hazardous impact on the environment and its removal is one the urgent need. The recent advancement have suggested microbial-based remediation is an efficient method of the removal of dyes, but unfortunately dye removal through the use of microbes still depends on the environmental changes. In this regard, many reported have suggested the proper oxygen transfer, operational stability, homogenization, less operational time, and suitability in hybrid bioreactors. The studies have provided the rationale for the application of microbial consortium of different microbes including bacteria, fungi, and algae. The microbial consortium has found to be more stress resistant with stability and enhanced adaptability over the single inoculation. The genome engineering of microbes may leads to the development of exceptionally adaptive techniques of bioremediation for high degradation of dyes. Hence, it is preferable to develop an alternative technology that could upgrade the microbial-based removal of dyes.
9. CONCLUSION
In times, bioremediation through microbes is gaining much important to because of its effectiveness, cost, and eco-friendly nature. Microbes have been also known to remediate the textile released dyes. Dyes and organic colored compounds are known to be hazardous pollutants which are categorized under xenobiotics. These are known to have several deleterious effects on aquatic and human life. Various types of microbes are known to remediate the dyes including fungi, bacteria, algae, and yeast. In the future, new microbial strains could be researched and could be used as bioremediation.
10. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
11. FUNDING
There is no funding to report.
12. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
13. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
14. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
15. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Saratale RG, Saratale GD, Chang JS, Govindwar SP. Decolorization and degradation of reactive azo dyes by fixed bed bioreactors containing immobilized cells of Proteus vulgaris NCIM-2027. Biotechnol Bioprocess Eng 2011;16:830. [CrossRef]
2. K?ížováH. Natural dyes:Their past, present, future and sustainability. In:Kanina, editor. Recent Developments in Fibrous Material Science. Prague:Kosmas Publishing;2015. 59-71.
3. Fleischmann C, Lievenbrück M, Ritter H. Polymers and dyes:Developments and applications. Polymers 2015;7:717-46. [CrossRef]
4. Sarkar S, Banerjee A, Halder U, Biswas R, Bandopadhyay R. Degradation of synthetic azo dyes of textile industry:A sustainable approach using microbial enzymes. Water Conserv Sci Eng 2017;2:121-31. [CrossRef]
5. Weiss B. Synthetic food colors and neurobehavioral hazards:The view from environmental health research. Environ Health Perspect 2012;120:1-5. [CrossRef]
6. Wainwright M. Dyes in the development of drugs and pharmaceuticals. Dyes Pigm 2008;76:582-9. [CrossRef]
7. Scarpi C, Ninci F, Centini M, Anselmi C. High-performance liquid chromatography determination of direct and temporary dyes in natural hair colourings. J Chromatogr A 1998;796:319-25. [CrossRef]
8. Kahn BE. The chemistry of photographic color dye formation. J Chem Educ 2004;81:694. [CrossRef]
9. Lodato A, Alfieri F, Olivieri G, Di Donato A, Marzocchella A, Salatino P. Azo-dye conversion by means of Pseudomonas sp. OX1. Enzyme Microb Technol 2007;41:646-52. [CrossRef]
10. Alabdraba WM, Bayati M. Biodegradation of azo dyes a review. Int J Environ Eng Nat Resour 2014;1:179-89.
11. McCurdy SM. Infrared processing of dry peas, canola, and canola screenings. J Food Sci 1992;57:941-4. [CrossRef]
12. I??k M, Sponza DT. Decolorization of azo dyes under batch anaerobic and sequential anaerobic/aerobic conditions. J Environ Sci Health A Tox Hazard Subst Environ Eng 2004;39:1107-27. [CrossRef]
13. Wong Y, Yu J. Laccase-catalyzed decolorization of synthetic dyes. Water Res 1999;33:3512-20. [CrossRef]
14. Carias CC, Novais JM, Martins-Dias S. Are Phragmites australis enzymes involved in the degradation of the textile azo dye acid orange 7?Bioresour Technol 2008;99:243-51. [CrossRef]
15. Nigam P, Banat IM, Singh D, Marchant R. Microbial process for the decolorization of textile effluent containing azo, diazo and reactive dyes. Process Biochem 1996;31:435-42. [CrossRef]
16. Banat IM, Nigam P, Singh D, Marchant R. Microbial decolorization of textile-dyecontaining effluents:A review. Bioresour Technol 1996;58:217-27. [CrossRef]
17. Lorimer JP, Mason TJ, Plattes M, Phull SS, Walton DJ. Degradation of dye effluent. Pure Appl Chem 2001;73:1957-68. [CrossRef]
18. Robinson T, McMullan G, Marchant R, Nigam P. Remediation of dyes in textile effluent:A critical review on current treatment technologies with a proposed alternative. Bioresour Technol 2001;77:247-55. [CrossRef]
19. Zhou W, Zimmermann W. Decolorization of industrial effluents containing reactive dyes by actinomycetes. FEMS Microbiol Lett 1993;107:157-61. [CrossRef]
20. Chander M, Arora DS. Evaluation of some white-rot fungi for their potential to decolourise industrial dyes. Dyes Pigm 2007;72:192-8. [CrossRef]
21. Kuhad RC, Sood N, Tripathi KK, Singh A, Ward OP. Developments in microbial methods for the treatment of dye effluents. Adv Appl Microbiol 2004;56:185-213. [CrossRef]
22. Saratale RG, Saratale GD, Chang JS, Govindwar SP. Decolorization and biodegradation of textile dye Navy blue HER by Trichosporon beigelii NCIM-3326. J Hazard Mater 2009;166:1421-8. [CrossRef]
23. Yusuf M, Shabbir M, Mohammad F. Natural colorants:Historical, processing and sustainable prospects. Nat Prod Bioprospect 2017;7:123-45. [CrossRef]
24. Serrano?Andrés L, Roos BO. A theoretical study of the indigoid dyes and their chromophore. Chem A Euro J 1997;3:717-25. [CrossRef]
25. Jones F, Kirby F. Naphthoquinone dyes in liquid crystaliine media. Mol Cryst Liq Cryst 1984;108:165-75. [CrossRef]
26. Sequin-Frey M. The chemistry of plant and animal dyes. J Chem Educ 1981;58:301. [CrossRef]
27. Vankar PS. Chemistry of natural dyes. Resonance 2000;5:73-80. [CrossRef]
28. Vettese Forster S, Christie RM. The significance of the introduction of synthetic dyes in the mid 19th century on the democratisation of western fashion. J Int Colour Assoc 2013;11:1-17.
29. Dos Santos AB, Cervantes FJ, van Lier JB. Review paper on current technologies for decolourisation of textile wastewaters:Perspectives for anaerobic biotechnology. Bioresour Technol 2007;98:2369-85. [CrossRef]
30. Baughman GL, Weber EJ. Transformation of dyes and related compounds in anoxic sediment:Kinetics and products. Environ Sci Technol 1994;28:267-76. [CrossRef]
31. Fontenot EJ, Lee YH, Matthews RD, Zhu G, Pavlostathis SG. Reductive decolorization of a textile reactive dyebath under methanogenic conditions. Appl Biochem Biotechnol 2003;109:207-25. [CrossRef]
32. Christie R. Environmental Aspects of Textile Dyeing. Boca Raton:Woodhead;2007. [CrossRef]
33. Hunger K. Dyes, general survey. In:Hunger K, editor. Industrial Dyes:Chemistry, Properties, Applications. Frankfurt:Wiley Subscription Services, Inc, A Wiley Company;2003. 1-10. [CrossRef]
34. Yang CL, McGarrahan J. Electrochemical coagulation for textile effluent decolorization. J Hazard Mater 2005;127:40-7. [CrossRef]
35. Correia VM, Stephenson T, Judd SJ. Characterisation of textile wastewaters?a review. Environ Technol 1994;15:917-29. [CrossRef]
36. Kalyani DC, Telke AA, Govindwar SP, Jadhav JP. Biodegradation and detoxification of reactive textile dye by isolated Pseudomonas sp. SUK1. Water Environ Res 2009;81:298-307. [CrossRef]
37. Jin XC, Liu GQ, Xu ZH, Tao WY. Decolorization of a dye industry effluent by Aspergillus fumigatus XC6. Appl Microbiol Biotechnol 2007;74:239-43. [CrossRef]
38. Chen KC, Wu JY, Liou DJ, Hwang SC. Decolorization of the textile dyes by newly isolated bacterial strains. J Biotechnol 2003;101:57-68. [CrossRef]
39. Hassaan MA, El Nemr A. Health and environmental impacts of dyes:Mini review. Am J Environ Sci Eng 2017;1:64-7.
40. Pandey A, Singh P, Iyengar L. Bacterial decolorization and degradation of azo dyes. Int Biodeterior Biodegrad 2007;59:73-84. [CrossRef]
41. Annuar MS, Adnan S, Vikineswary S, Chisti Y. Kinetics and energetics of azo dye decolorization by Pycnoporus sanguineus. Water Air Soil Pollut 2009;202:179-88. [CrossRef]
42. Zhao X, Hardin IR. HPLC and spectrophotometric analysis of biodegradation of azo dyes by Pleurotus ostreatus. Dyes Pigm 2007;73:322-5. [CrossRef]
43. Kumar PS, Narayan AS, Dutta A. Nanochemicals and effluent treatment in textile industries. In:Muthu SS, editor. Textiles and Clothing Sustainability:Nanotextiles and Sustainability. Singapore:Springer Singapore;2017. 57-96. [CrossRef]
44. Akarslan F, Demiralay H. Effects of textile materials harmful to human health. Acta Phys Pol A 2015;128:407-9. [CrossRef]
45. Singh RL, Singh PK, Singh RP. Enzymatic decolorization and degradation of azo dyes-a review. Int Biodeterior Biodegrad 2015;104:21-31. [CrossRef]
46. Gomathi E, Rathika G, Elango S. Physico-chemical parameters of textile dyeing effluent and its impacts with casestudy. Int J Res Chem Environ 2017;7:17-24.
47. Adeyemo AA, Adeoye IO, Bello OS. Adsorption of dyes using different types of clay:A review. Appl Water Sci 2017;7:543-68. [CrossRef]
48. Foo K, Hameed BH. An overview of dye removal via activated carbon adsorption process. Desalin Water Treat 2010;19:255-74. [CrossRef]
49. Zaharia C, Suteu D, Muresan A. Options and solutions for textile effluent decolorization using some specific physico-chemical treatment steps. Environ Eng Manag J 2012;11:493-509. [CrossRef]
50. Sahu O, Singh N. 13-Significance of bioadsorption process on textile industry wastewater. In:Shahid Ul I, Butola BS, editors. The Impact and Prospects of Green Chemistry for Textile Technology. Sawston, Cambridge:Woodhead Publishing;2019. 367-416. [CrossRef]
51. Adegoke KA, Bello OS. Dye sequestration using agricultural wastes as adsorbents. Water Resour Ind 2015;12:8-24. [CrossRef]
52. Mane VP, Patil SS, Syed AA, Baig MM. Bioconversion of low quality lignocellulosic agricultural waste into edible protein by Pleurotus sajor-caju (Fr.) Singer. J Zhejiang Univ Sci 2007;8:745-51. [CrossRef]
53. Shah MP, Patel KA, Darji A. Microbial degradation and decolorization of methyl orange dye by an application of Pseudomonas spp. ETL-1982. Int J Environ Bioremediat Biodegrad 2013;1:26-36.
54. Joshi M, Bansal R, Purwar R. Colour removal from textile effluents. Indian J Fibre Text Res 2004;29:239-59.
55. Kim S, Park C, Kim TH, Lee J, Kim SW. COD reduction and decolorization of textile effluent using a combined process. J Biosci Bioeng 2003;95:102-5. [CrossRef]
56. Ghanbari F, Moradi M, Eslami A, Emamjomeh MM. Electrocoagulation/flotation of textile wastewater with simultaneous application of aluminum and iron as anode. Environ Process 2014;1:447-57. [CrossRef]
57. Getoff N, Lutz W. Radiation induced decomposition of hydrocarbons in water resources. Radiat Phys Chem 1985;25:21-6. [CrossRef]
58. Kuo W. Decolorizing dye wastewater with Fenton's reagent. Water Res 1992;26:881-6. [CrossRef]
59. Kiran S, Ali S, Asgher M, Shahid SA. Photo-fenton process:Optimization and decolourization and mineralization of reactive blue 222 dye. J Environ Sci Water Resour 2012;1:267.
60. Yoon Y, Hwang Y, Kwon M, Jung Y, Hwang TM, Kang JW. Application of O3 and O3/H2O2 as post-treatment processes for color removal in swine wastewater from a membrane filtration system. J Ind Eng Chem 2014;20:2801-5. [CrossRef]
61. Palit S. An overview of ozonation associated with nanofiltration as an effective procedure in treating dye effluents from textile industries with the help of a bubble column reactor. Int J Chem Sci 2012;10:27-35.
62. De Brito-Pelegrini N, De Tarso Ferreira Sales P, Pelegrini R. Photochemical treatment of industrial textile effluent containing reactive dyes. Environ Technol 2007;28:321-8. [CrossRef]
63. Azbar N, Yonar T, Kestioglu K. Comparison of various advanced oxidation processes and chemical treatment methods for COD and color removal from a polyester and acetate fiber dyeing effluent. Chemosphere 2004;55:35-43. [CrossRef]
64. Slokar YM, Le Marechal AM. Methods of decoloration of textile wastewaters. Dyes Pigm 1998;37:335-56. [CrossRef]
65. Pignata C, Fea E, Rovere R, Degan R, Lorenzi E, de Ceglia M, et al. Chlorination in a wastewater treatment plant:Acute toxicity effects of the effluent and of the recipient water body. Environ Monit Assess 2012;184:2091-103. [CrossRef]
66. Parsa JB, Abbasi M. Decolorization of synthetic and real wastewater by indirect electrochemical oxidation process. Acta Chim Slov 2007;54:792-6.
67. Kaushik P, Malik A. Fungal dye decolourization:Recent advances and future potential. Environ Int 2009;35:127-41. [CrossRef]
68. Rodríguez Couto S. Dye removal by immobilised fungi. Biotechnol Adv 2009;27:227-35. [CrossRef]
69. Ahmad A, Mohd-Setapar SH, Chuong CS, Khatoon A, Wani WA, Kumar R, et al. Recent advances in new generation dye removal technologies:Novel search for approaches to reprocess wastewater. RSC Adv 2015;5:30801-18. [CrossRef]
70. Prabha S, Gogoi A, Mazumder P, Ramanathan AL, Kumar M. Assessment of the impact of textile effluents on microbial diversity in Tirupur district, Tamil Nadu. Appl Water Sci 2017;7:2267-77. [CrossRef]
71. Parmar ND, Shukla SR. Decolourization of dye wastewater by microbial methods-a review. Indian J Chem Technol 2018;25:315-23.
72. Gill PK, Arora DS, Chander M. Biodecolourization of azo and triphenylmethane dyes by Dichomitus squalens and Phlebia spp. J Ind Microbiol Biotechnol 2002;28:201-3. [CrossRef]
73. SvobodováK, Senholdt M, Novotný?, Rehorek A. Mechanism of reactive orange 16 degradation with the white rot fungus Irpex lacteus. Process Biochem 2007;42:1279-84. [CrossRef]
74. Jayasinghe C, Imtiaj A, Lee GW, Im KH, Hur H, Lee MW, et al. Degradation of three aromatic dyes by white rot fungi and the production of ligninolytic enzymes. Mycobiology 2008;36:114-20. [CrossRef]
75. Abd El-Zaher EH. Biodegradation of reactive dyes using soil fungal isolates and Ganoderma resinaceum. Ann Microbiol 2010;60:269-78. [CrossRef]
76. Gomi N, Yoshida S, Matsumoto K, Okudomi M, Konno H, Hisabori T, et al. Degradation of the synthetic dye amaranth by the fungus Bjerkandera adusta Dec 1:Inference of the degradation pathway from an analysis of decolorized products. Biodegradation 2011;22:1239-45. [CrossRef]
77. Hadibarata T, Yusoff AR, Aris A, Salmiati S, Hidayat T, Kristanti RA. Decolorization of azo, triphenylmethane and anthraquinone dyes by laccase of a newly isolated Armillaria sp. F022. Water Air Soil Pollut 2012;223:1045-54. [CrossRef]
78. Chakraborty S, Basak B, Dutta S, Bhunia B, Dey A. Decolorization and biodegradation of congo red dye by a novel white rot fungus Alternaria alternata CMERI F6. Bioresour Technol 2013;147:662-6. [CrossRef]
79. Ma L, Zhuo R, Liu H, Yu D, Jiang M, Zhang X, et al. Efficient decolorization and detoxification of the sulfonated azo dye Reactive Orange 16 and simulated textile wastewater containing Reactive Orange 16 by the white-rot fungus Ganoderma sp. En3 isolated from the forest of Tzu-chin Mountain in China. Biochem Eng J 2014;82:1-9. [CrossRef]
80. Chen SH, Ting AS. Biodecolorization and biodegradation potential of recalcitrant triphenylmethane dyes by Coriolopsis sp. isolated from compost. J Environ Manage 2015;150:274-80. [CrossRef]
81. Kristanti RA, Zubir MM, Hadibarata T. Biotransformation studies of cresol red by Absidia spinosa M15. J Environ Manage 2016;172:107-11. [CrossRef]
82. Barapatre A, Aadil KR, Jha H. Biodegradation of malachite green by the ligninolytic fungus Aspergillus flavus. Clean Soil Air Water 2017;45:1600045. [CrossRef]
83. Khan R, Fulekar MH. Mineralization of a sulfonated textile dye Reactive Red 31 from simulated wastewater using pellets of Aspergillus bombycis. Bioresour Bioprocess 2017;4:23. [CrossRef]
84. Asses N, Ayed L, Hkiri N, Hamdi M. Congo red decolorization and detoxification by Aspergillus niger:Removal mechanisms and dye degradation pathway. BioMed Res Int 2018;2018:1-9. [CrossRef]
85. Iark D, Buzzo AJ, Garcia JA, Côrrea VG, Helm CV, Corrêa RC, et al. Enzymatic degradation and detoxification of azo dye Congo red by a new laccase from Oudemansiella canarii. Bioresour Technol 2019;289:121655. [CrossRef]
86. Singh G, Dwivedi SK. Decolorization and degradation of Direct Blue-1 (Azo dye) by newly isolated fungus Aspergillus terreus GS28, from sludge of carpet industry. Environ Technol Innov 2020;18:100751. [CrossRef]
87. Krishnan S, Zulkapli NS, Din MF, Majid ZA, Nasrullah M, Sairan FM. Photocatalytic degradation of methylene blue dye and fungi Fusarium equiseti using titanium dioxide recovered from drinking water treatment sludge. In:Biomass Conversion and Biorefinery. Berlin:Springer;2021. [CrossRef]
88. Kalyani DC, Telke AA, Dhanve RS, Jadhav JP. Ecofriendly biodegradation and detoxification of Reactive Red 2 textile dye by newly isolated Pseudomonas sp. SUK1. J Hazard Mater 2009;163:735-42. [CrossRef]
89. Ali H. Biodegradation of synthetic dyes-a review. Water Air Soil Pollut 2010;213:251-73. [CrossRef]
90. Saratale RG, Saratale GD, Chang JS, Govindwar SP. Bacterial decolorization and degradation of azo dyes:A review. J Taiwan Inst Chem Eng 2011;42:138-57. [CrossRef]
91. Kapdan IK, Oztekin R. Decolorization of textile dyestuff Reactive Orange 16 in fed-batch reactor under anaerobic condition. Enzyme Microb Technol 2003;33:231-5. [CrossRef]
92. Park EH, Jang MS, Cha IH, Choi YL, Cho YS, Kim CH, et al. Decolorization of a sulfonated azo dye, Congo Red, by Staphylococcus sp. EY-3. J Microbiol Biotechnol 2005;15:221-5.
93. Kalyani DC, Patil PS, Jadhav JP, Govindwar SP. Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp. SUK1. Bioresour Technol 2008;99:4635-41. [CrossRef]
94. Parshetti GK, Telke AA, Kalyani DC, Govindwar SP. Decolorization and detoxification of sulfonated azo dye methyl orange by Kocuria rosea MTCC 1532. J Hazard Mater 2010;176:503-9. [CrossRef]
95. Sugumar S, Thangam E. Biodegradation and decolorization of Reactive orange 16 by Nocardiopsis alba soil isolate. J Bioremediat Biodegrad 2012;3:155.
96. Ghanem KM, Al-Fassi FA, Biag AK. Optimization of methyl orange decolorization by mono and mixed bacterial culture techniques using statistical designs. Afr J Microbiol Res 2012;6:436-46. [CrossRef]
97. Shah MP, Patel KA, Nair SS, Darji AM, Maharaul S. Exploited application of Bacillus spp. ETL-1979 for degradation and decolorization of Methyl orange, Malachite green and Congo red. J Bioremed Biodeg 2013;4:1-6.
98. Cui D, Li G, Zhao M, Han S. Decolourization of azo dyes by a newly isolated Klebsiella sp. strain Y3, and effects of various factors on biodegradation. Biotechnol Biotechnol Equip 2014;28:478-86. [CrossRef]
99. Ng IS, Chen T, Lin R, Zhang X, Ni C, Sun D. Decolorization of textile azo dye and Congo red by an isolated strain of the dissimilatory manganese-reducing bacterium Shewanella xiamenensis BC01. Appl Microbiol Biotechnol 2014;98:2297-308. [CrossRef]
100. Babu SS, Mohandass C, Vijayaraj AS, Dhale MA. Detoxification and color removal of Congo red by a novel Dietzia sp. (DTS26)-a microcosm approach. Ecotoxicol Environ Saf 2015;114:52-60. [CrossRef]
101. Roat C, Kadam A, Patel T, Dave S. Biodegradation of diazo dye, reactive blue 160 by isolate Microbacterium sp. B12 mutant:Identification of intermediates by LC-MS. Int J Curr Microbiol Appl Sci 2016;5:534-47. [CrossRef]
102. Abo-State MA, Saleh YE, Hazaa H. Decolorization of Congo Red dye by bacterial isolates. J Ecol Health Environ 2017;5:41-8.
103. Ajaz M, Elahi A, Rehman A. Degradation of azo dye by bacterium, Alishewanella sp. CBL-2 isolated from industrial effluent and its potential use in decontamination of wastewater. J Water Reuse Desalin 2018;8:507-15. [CrossRef]
104. Mani P, Fidal VT, Bowman K, Breheny M, Chandra TS, Keshavarz T, et al. Degradation of azo dye (acid orange 7) in a microbial fuel cell:Comparison between anodic microbial-mediated reduction and cathodic laccase-mediated oxidation. Front Energ Res 2019;7:101. [CrossRef]
105. Sari IP, Simarani K. Comparative static and shaking culture of metabolite derived from methyl red degradation by Lysinibacillus fusiformis strain W1B6. R Soc Open Sci 2019;6:190152. [CrossRef]
106. Saha P, Rao KV. Biotransformation of reactive orange 16 by alkaliphilic bacterium Bacillus flexus VITSP6 and toxicity assessment of biotransformed metabolites. Int J Environ Sci Technol 2020;17:99-114. [CrossRef]
107. Prabhakar Y, Gupta A, Kaushik A. Microbial degradation of reactive red-35 dye:Upgraded progression through Box-Behnken design modeling and cyclic acclimatization. J Water Process Eng 2021;40:101782. [CrossRef]
108. Akansha K, Yadav AN, Kumar M, Chakraborty D, Sachan SG. Decolorization and degradation of reactive orange 16 by Bacillus stratosphericus SCA1007. Folia Microbiol 2022;67:91-102. [CrossRef]
109. Yu Z, Wen X. Screening and identification of yeasts for decolorizing synthetic dyes in industrial wastewater. Int Biodeterior Biodegrad 2005;56:109-14. [CrossRef]
110. Deivasigamani C, Das N. Biodegradation of basic violet 3 by Candida krusei isolated from textile wastewater. Biodegradation 2011;22:1169-80. [CrossRef]
111. Qu Y, Cao X, Ma Q, Shi S, Tan L, Li X, et al. Aerobic decolorization and degradation of Acid Red B by a newly isolated Pichia sp. TCL. J Hazard Mater 2012;223-224:31-8. [CrossRef]
112. Tan L, Ning S, Zhang X, Shi S. Aerobic decolorization and degradation of azo dyes by growing cells of a newly isolated yeast Candida tropicalis TL-F1. Bioresour Technol 2013;138:307-13. [CrossRef]
113. Tan L, Li H, Ning S, Xu B. Aerobic decolorization and degradation of azo dyes by suspended growing cells and immobilized cells of a newly isolated yeast Magnusiomyces ingens LH-F1. Bioresour Technol 2014;158:321-8. [CrossRef]
114. Li H, Tan L, Ning S, He M. Reactor performance and microbial community dynamics during aerobic degradation and detoxification of Acid Red B with activated sludge bioaugmented by a yeast Candida tropicalis TL-F1 in MBR. Int Biodeterior Biodegrad 2015;104:149-56. [CrossRef]
115. Tan L, He M, Song L, Fu X, Shi S. Aerobic decolorization, degradation and detoxification of azo dyes by a newly isolated salt-tolerant yeast Scheffersomyces spartinae TLHS-SF1. Bioresour Technol 2016;203:287-94. [CrossRef]
116. Rosu CM, Avadanei M, Gherghel D, Mihasan M, Mihai C, Trifan A, et al. Biodegradation and detoxification efficiency of azo-dye Reactive Orange 16 by Pichia kudriavzevii CR-Y103. Water Air Soil Pollut 2017;229:15. [CrossRef]
117. Song Z, Song L, Shao Y, Tan L. Degradation and detoxification of azo dyes by a salt-tolerant yeast Cyberlindnera samutprakarnensis S4 under high-salt conditions. World J Microbiol Biotechnol 2018;34:131. [CrossRef]
118. Guo G, Tian F, Zhao Y, Tang M, Liu W, Liu C, et al. Aerobic decolorization and detoxification of Acid Scarlet GR by a newly isolated salt-tolerant yeast strain Galactomyces geotrichum GG. Int Biodeterior Biodegrad 2019;145:104818. [CrossRef]
119. Al-Tohamy R, Kenawy ER, Sun J, Ali SS. Performance of a newly isolated salt-tolerant yeast strain Sterigmatomyces halophilus SSA-1575 for azo dye decolorization and detoxification. Front Microbiol 2020;11:1163. [CrossRef]
120. Wang Y, Xu B, Ning S, Shi S, Tan L. Magnetically stimulated azo dye biodegradation by a newly isolated osmo-tolerant Candida tropicalis A1 and transcriptomic responses. Ecotoxicol Environ Saf 2021;209:111791. [CrossRef]
121. Semple KT, Cain RB, Schmidt S. Biodegradation of aromatic compounds by microalgae. FEMS Microbiol Lett 1999;170:291-300. [CrossRef]
122. Parikh A, Madamwar D. Textile dye decolorization using Cyanobacteria. Biotechnol Lett 2005;27:323-6. [CrossRef]
123. Omar HH. Algal decolorization and degradation of monoazo and diazo dyes. Pak J Biol Sci 2008;11:1310-6. [CrossRef]
124. Khataee AR, Dehghan G, Ebadi A, Zarei M, Pourhassan M. Biological treatment of a dye solution by Macroalgae Chara sp.:Effect of operational parameters, intermediates identification and artificial neural network modeling. Bioresour Technol 2010;101:2252-8. [CrossRef]
125. Al-Fawwaz A, Jacob JH. Removal of methylene blue and malachite green from aqueous solutions by Chlorella and Chlamydomonas species isolated from a thermal spring environment. Int J Integr Biol 2011;12:36-40.
126. Sarwa P, Verma SK. Decolourization of Orange G Dye by microalgae Acutodesmus obliquues strain PSV2 isolated from textile industrial site. Int J Appl Sci Biotechnol 2013;1:247-52. [CrossRef]
127. Chia MA, Odoh OA, Ladan Z. The indigo blue dye decolorization potential of immobilized Scenedesmus quadricauda. Water Air Soil Pollut 2014;225:1920. [CrossRef]
128. Mahalakshmi S, Lakshmi D, Menaga U. Biodegradation of different concentration of dye (Congo red dye) by using green and blue green algae. Int J Environ Res 2015;9:735-44.
129. Al-Fawwaz AT, Abdullah M. Decolorization of methylene blue and malachite green by immobilized Desmodesmus sp. isolated from North Jordan. Int J Environ Sci Dev 2016;7:95-9. [CrossRef]
130. Revathi S, Kumar SM, Santhanam P, Kumar SD, Son N, Kim MK. Bioremoval of the indigo blue dye by immobilized microalga Chlorella vulgaris (PSBDU06). J Sci Ind Res 2017;76:50-6.
131. Khehra MS, Saini HS, Sharma DK, Chadha BS, Chimni SS. Comparative studies on potential of consortium and constituent pure bacterial isolates to decolorize azo dyes. Water Res 2005;39:5135-41. [CrossRef]
132. Forgacs E, Cserháti T, Oros G. Removal of synthetic dyes from wastewaters:A review. Environ Int 2004;30:953-71. [CrossRef]
133. Nigam P, Mc Mullan G, Banat IM, Marchant R. Decolourisation of effluent from the textile industry by a microbial consortium. Biotechnol Lett 1996;18:117-20. [CrossRef]
134. Tony BD, Goyal D, Khanna S. Decolorization of textile azo dyes by aerobic bacterial consortium. Int Biodeterior Biodegr 2009;63:462-9. [CrossRef]
135. Patil PS, Phugare SS, Jadhav SB, Jadhav JP. Communal action of microbial cultures for Red HE3B degradation. J Hazard Mater 2010;181:263-70. [CrossRef]
136. Waghmode TR, Kurade MB, Khandare RV, Govindwar SP. A sequential aerobic/microaerophilic decolorization of sulfonated mono azo dye Golden Yellow HER by microbial consortium GG-BL. Int Biodeterior Biodegrad 2011;65:1024-34. [CrossRef]
137. Cui D, Li G, Zhao D, Gu X, Wang C, Zhao M. Microbial community structures in mixed bacterial consortia for azo dye treatment under aerobic and anaerobic conditions. J Hazard Mater 2012;221-222:185-92. [CrossRef]
138. Karunya A, Nachiyar CV, Ananth PB, Sunkar S, Jabasingh SA. Development of microbial consortium CN-1 for the degradation of Mordant Black 17. J Environ Chem Eng 2014;2:832-40. [CrossRef]
139. Lade H, Kadam A, Paul D, Govindwar S. Biodegradation and detoxification of textile azo dyes by bacterial consortium under sequential microaerophilic/aerobic processes. EXCLI J 2015;14:158-74.
140. Holey BA. Decolourization of Congo Red dye by bacteria and consortium isolated from dye contaminated soil. Int Res J Sci Eng 2015;3:107-12.
141. Mahmood R, Sharif F, Ali S, Hayyat MU. Enhancing the decolorizing and degradation ability of bacterial consortium isolated from textile effluent affected area and its application on seed germination. Sci World J 2015;2015:1-10. [CrossRef]
142. Kuppusamy S, Sethurajan M, Kadarkarai M, Aruliah R. Biodecolourization of textile dyes by novel, indigenous Pseudomonas stutzeri MN1 and Acinetobacter baumannii MN3. J Environ Chem Eng 2017;5:716-24. [CrossRef]
143. Karim ME, Dhar K, Hossain MT. Decolorization of textile reactive dyes by bacterial monoculture and consortium screened from textile dyeing effluent. J Genet Eng Biotechnol 2018;16:375-80. [CrossRef]
144. Ali SS, Al-Tohamy R, Xie R, El-Sheekh MM, Sun J. Construction of a new lipase-and xylanase-producing oleaginous yeast consortium capable of reactive azo dye degradation and detoxification. Bioresour Technol 2020;313:123631. [CrossRef]
145. Guo G, Li X, Tian F, Liu T, Yang F, Ding K, et al. Azo dye decolorization by a halotolerant consortium under microaerophilic conditions. Chemosphere 2020;244:125510. [CrossRef]
146. El-Sheekh MM, El-Shanshoury AR, Abou-El-Souod GW, Gharieb DY, El Shafay SM. Decolorization of dyestuffs by some species of green algae and Cyanobacteria and its consortium. Int J Environ Sci Technol 2021;18:3895-906. [CrossRef]
147. Phugare SS, Kalyani DC, Patil AV, Jadhav JP. Textile dye degradation by bacterial consortium and subsequent toxicological analysis of dye and dye metabolites using cytotoxicity, genotoxicity and oxidative stress studies. J Hazard Mater 2011;186:713-23. [CrossRef]
148. Chen Y, Zhang L, Feng L, Chen G, Wang Y, Zhai Z, et al. Exploration of the key functional strains from an azo dye degradation microbial community by DGGE and high-throughput sequencing technology. Environ Sci Pollut Res Int 2019;26:24658-71. [CrossRef]
149. Thiruppathi K, Rangasamy K, Ramasamy M, Muthu D. Evaluation of textile dye degrading potential of ligninolytic bacterial consortia. Environ Chall 2021;4:100078. [CrossRef]
150. Mohanty SS, Kumar A. Enhanced degradation of anthraquinone dyes by microbial monoculture and developed consortium through the production of specific enzymes. Sci Rep 2021;11:7678. [CrossRef]
151. Jain K, Shah V, Chapla D, Madamwar D. Decolorization and degradation of azo dye-Reactive Violet 5R by an acclimatized indigenous bacterial mixed cultures-SB4 isolated from anthropogenic dye contaminated soil. J Hazard Mater 2012;213-214:378-86. [CrossRef]
152. Jadhav SU, Jadhav MU, Kagalkar AN, Govindwar SP. Decolorization of brilliant Blue G dye mediated by degradation of the microbial consortium of Galactomyces geotrichum and Bacillus sp. J Chin Inst Chem Eng 2008;39:563-70. [CrossRef]
153. Khehra MS, Saini HS, Sharma DK, Chadha BS, Chimni SS. Decolorization of various azo dyes by bacterial consortium. Dyes Pigm 2005;67:55-61. [CrossRef]
154. Joshi T, Iyengar L, Singh K, Garg S. Isolation, identification and application of novel bacterial consortium TJ-1 for the decolourization of structurally different azo dyes. Bioresour Technol 2008;99:7115-21. [CrossRef]
155. Patel Y, Mehta C, Gupte A. Assessment of biological decolorization and degradation of sulfonated di-azo dye Acid Maroon V by isolated bacterial consortium EDPA. Int Biodeterior Biodegrad 2012;75:187-93. [CrossRef]
156. Eskandari F, Shahnavaz B, Mashreghi M. Optimization of complete RB-5 azo dye decolorization using novel cold-adapted and mesophilic bacterial consortia. J Environ Manage 2019;241:91-8. [CrossRef]
157. Mohana S, Shrivastava S, Divecha J, Madamwar D. Response surface methodology for optimization of medium for decolorization of textile dye direct black 22 by a novel bacterial consortium. Bioresour Technol 2008;99:562-9. [CrossRef]
158. Lade HS, Waghmode TR, Kadam AA, Govindwar SP. Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int Biodeterior Biodegrad 2012;72:94-107. [CrossRef]
159. Dafale N, Rao NN, Meshram SU, Wate SR. Decolorization of azo dyes and simulated dye bath wastewater using acclimatized microbial consortium-Biostimulation and halo tolerance. Bioresour Technol 2008;99:2552-8. [CrossRef]
160. Dafale N, Wate S, Meshram S, Nandy T. Kinetic study approach of remazol black-B use for the development of two-stage anoxic-oxic reactor for decolorization/biodegradation of azo dyes by activated bacterial consortium. J Hazard Mater 2008;159:319-28. [CrossRef]
161. Eslami H, Shariatifar A, Rafiee E, Shiranian M, Salehi F, Hosseini SS, et al. Decolorization and biodegradation of reactive Red 198 Azo dye by a new Enterococcus faecalis-Klebsiella variicola bacterial consortium isolated from textile wastewater sludge. World J Microbiol Biotechnol 2019;35:38. [CrossRef]
162. Kumar K, Devi SS, Krishnamurthi K, Dutta D, Chakrabarti T. Decolorisation and detoxification of Direct Blue-15 by a bacterial consortium. Bioresour Technol 2007;98:3168-71. [CrossRef]
163. Deive FJ, Domínguez A, Barrio T, Moscoso F, Morán P, Longo MA, et al. Decolorization of dye Reactive Black 5 by newly isolated thermophilic microorganisms from geothermal sites in Galicia (Spain). J Hazard Mater 2010;182:735-42. [CrossRef]
164. Krishnamoorthy R, Jose PA, Ranjith M, Anandham R, Suganya K, Prabhakaran J, et al. Decolourisation and degradation of azo dyes by mixed fungal culture consisted of Dichotomomyces cejpii MRCH 1-2 and Phoma tropica MRCH 1-3. J Environ Chem Eng 2018;6:588-95. [CrossRef]
165. Shah MP, Bera SP. Microbial treatment of textile dye reactive Red 3 by a newly developed bacterial consortium. Nanotechnol Environ Eng 2021;6:62. [CrossRef]
166. Manogaran M, Yasid NA, Othman AR, Gunasekaran B, Halmi MI, Shukor MY. Biodecolourisation of reactive Red 120 as a sole carbon source by a bacterial consortium-toxicity assessment and statistical optimisation. Int J Environ Res Public Health 2021;18:2424. [CrossRef]
167. Gou M, Qu Y, Zhou J, Ma F, Tan L. Azo dye decolorization by a new fungal isolate, Penicillium sp. QQ and fungal-bacterial cocultures. J Hazard Mater 2009;170:314-9. [CrossRef]
168. Xu JZ, Zhang JL, Hu KH, Zhang WG. The relationship between lignin peroxidase and manganese peroxidase production capacities and cultivation periods of mushrooms. Microbial Biotechnol 2013;6:241-7. [CrossRef]
169. Shaffiqu TS, Roy JJ, Nair RA, Abraham TE. Degradation of textile dyes mediated by plant peroxidases. Appl Biochem Biotechnol 2002;102-103:315-26. [CrossRef]
170. Peralta-Zamora P, de Moraes SG, Pelegrini R, Freire M, Reyes J, Mansilla H, et al. Evaluation of ZnO, TiO2 and supported ZnO on the photoassisted remediation of black liquor, cellulose and textile mill effluents. Chemosphere 1998;36:2119-33. [CrossRef]
171. Pramanik S, Chaudhuri S. Laccase activity and azo dye decolorization potential of Podoscypha elegans. Mycobiology 2018;46:79-83. [CrossRef]
172. Kalyani D, Dhiman SS, Kim H, Jeya M, Kim IW, Lee JK. Characterization of a novel laccase from the isolated Coltricia perennis and its application to detoxification of biomass. Process Biochem 2012;47:671-8. [CrossRef]
173. Solano F, Garcia E, Perez D, Sanchez-Amat A. Isolation and characterization of strain MMB-1 (CECT 4803), a novel melanogenic marine bacterium. Appl Environ Microbiol 1997;63:3499-506. [CrossRef]
174. Ridge JP, Lin M, Larsen EI, Fegan M, McEwan AG, Sly LI. A multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ Microbiol 2007;9:944-53. [CrossRef]
175. Kalme S, Jadhav S, Jadhav M, Govindwar S. Textile dye degrading laccase from Pseudomonas desmolyticum NCIM 2112. Enzyme Microb Technol 2009;44:65-71. [CrossRef]
176. Telke AA, Ghodake GS, Kalyani DC, Dhanve RS, Govindwar SP. Biochemical characteristics of a textile dye degrading extracellular laccase from a Bacillus sp. ADR. Bioresour Technol 2011;102:1752-6. [CrossRef]
177. Forootanfar H, Moezzi A, Aghaie-Khozani M, Mahmoudjanlou Y, Ameri A, Niknejad F, et al. Synthetic dye decolorization by three sources of fungal laccase. Iran J Environ Health Sci Eng 2012;9:27. [CrossRef]
178. Solís M, Solís A, Pérez HI, Manjarrez N, Flores M. Microbial decolouration of azo dyes:A review. Process Biochem 2012;47:1723-48. [CrossRef]
179. Russ R, Rau J, Stolz A. The function of cytoplasmic flavin reductases in the reduction of azo dyes by bacteria. Appl Environ Microbiol 2000;66:1429-34. [CrossRef]
180. Chen H, Hopper SL, Cerniglia CE. Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoprotein. Microbiology 2005;151:1433-41. [CrossRef]
181. Leelakriangsak M, Borisut S. Characterization of the decolorizing activity of azo dyes by Bacillus subtilis azoreductase AzoR1. Songklanakarin J Sci Technol 2012;34:509-16.
182. Elfarash A, Mawad AM, Yousef NM, Shoreit AA. Azoreductase kinetics and gene expression in the synthetic dyes-degrading Pseudomonas. Egypt J Basic Appl Sci 2017;4:315-22. [CrossRef]
183. Agrawal S, Tipre D, Patel B, Dave S. Optimization of triazo acid black 210 dye degradation by Providencia sp. SRS82 and elucidation of degradation pathway. Process Biochem 2014;49:110-9. [CrossRef]
184. Durán N, Esposito E. Potential applications of oxidative enzymes and phenoloxidase-like compounds in wastewater and soil treatment:A review. Appl Catal B Environ 2000;28:83-99. [CrossRef]
185. Tien M, Kirk TK, Bull C, Fee JA. Steady-state and transient-state kinetic studies on the oxidation of 3,4-dimethoxybenzyl alcohol catalyzed by the ligninase of Phanerocheate chrysosporium Burds. J Biol Chem 1986;261:1687-93. [CrossRef]
186. Glenn JK, Akileswaran L, Gold MH. Mn(II) oxidation is the principal function of the extracellular Mn-peroxidase from Phanerochaete chrysosporium. Arch Biochem Biophys 1986;251:688-96. [CrossRef]
187. Glenn JK, Morgan MA, Mayfield MB, Kuwahara M, Gold MH. An extracellular H2O2-requiring enzyme preparation involved in lignin biodegradation by the white rot basidiomycete Phanerochaete chrysosporium. Biochem Biophys Res Commun 1983;114:1077-83. [CrossRef]
188. Roberts JN, Singh R, Grigg JC, Murphy ME, Bugg TD, Eltis LD. Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry 2011;50:5108-19. [CrossRef]
189. Salvachúa D, Prieto A, Martínez ÁT, Martínez MJ. Characterization of a novel dye-decolorizing peroxidase (DyP)-type enzyme from Irpex lacteus and its application in enzymatic hydrolysis of wheat straw. Appl Environ Microbiol 2013;79:4316-24. [CrossRef]
190. Chen C, Shrestha R, Jia K, Gao PF, Geisbrecht BV, Bossmann SH, et al. Characterization of dye-decolorizing peroxidase (DyP) from Thermomonospora curvata reveals unique catalytic properties of A-type DyPs. J Biol Chem 2015;290:23447-63. [CrossRef]
191. Min K, Gong G, Woo HM, Kim Y, Um Y. A dye-decolorizing peroxidase from Bacillus subtilis exhibiting substrate-dependent optimum temperature for dyes and b-ether lignin dimer. Sci Rep 2015;5:8245. [CrossRef]
192. Shrestha R, Huang G, Meekins DA, Geisbrecht BV, Li P. Mechanistic insights into dye-decolorizing peroxidase revealed by solvent isotope and viscosity effects. ACS Catal 2017;7:6352-64. [CrossRef]
193. Ogugbue CJ, Sawidis T. Bioremediation and detoxification of synthetic wastewater containing triarylmethane dyes by Aeromonas hydrophila isolated from industrial effluent. Biotechnol Res Int 2011;2011:967925. [CrossRef]
194. Nozaki K, Beh CH, Mizuno M, Isobe T, Shiroishi M, Kanda T, et al. Screening and investigation of dye decolorization activities of Basidiomycetes. J Biosci Bioeng 2008;105:69-72. [CrossRef]
195. Wang H, Zheng XW, Su JQ, Tian Y, Xiong XJ, Zheng TL. Biological decolorization of the reactive dyes Reactive Black 5 by a novel isolated bacterial strain Enterobacter sp. EC3. J Hazard Mater 2009;171:654-9. [CrossRef]
196. Esmaeili A, Kalantari M. Bioremoval of an azo textile dye, reactive Red 198, by Aspergillus flavus. World J Microbiol Biotechnol 2012;28:1125-31. [CrossRef]
197. Saratale RG, Gandhi SS, Purankar MV, Kurade MB, Govindwar SP, Oh SE, et al. Decolorization and detoxification of sulfonated azo dye C.I. remazol red and textile effluent by isolated Lysinibacillus sp. RGS. J Biosci Bioeng 2013;115:658-67. [CrossRef]
198. Khan Z, Jain K, Soni A, Madamwar D. Microaerophilic degradation of sulphonated azo dye-reactive red 195 by bacterial consortium AR1 through co-metabolism. Int Biodeterior Biodegrad 2014;94:167-75. [CrossRef]
199. Huang G, Wang W, Liu G. Simultaneous chromate reduction and azo dye decolourization by Lactobacillus paracase CL1107 isolated from deep sea sediment. J Environ Manage 2015;157:297-302. [CrossRef]
200. Ali HM, Shehata SF, Ramadan KM. Microbial decolorization and degradation of crystal violet dye by Aspergillus niger. Int J Environ Sci Technol 2016;13:2917-26. [CrossRef]
201. Guadie A, Tizazu S, Melese M, Guo W, Ngo HH, Xia S. Biodecolorization of textile azo dye using Bacillus sp. strain CH12 isolated from alkaline lake. Biotechnol Rep (Amst) 2017;15:92-100. [CrossRef]
202. Roy DC, Biswas SK, Saha AK, Sikdar B, Rahman M, Roy AK, et al. Biodegradation of crystal violet dye by bacteria isolated from textile industry effluents. PeerJ 2018;6:e5015. [CrossRef]
203. Hossen MZ, Hussain ME, Hakim A, Islam K, Uddin MN, Azad AK. Biodegradation of reactive textile dye Novacron Super Black G by free cells of newly isolated Alcaligenes faecalis AZ26 and Bacillus spp obtained from textile effluents. Heliyon 2019;5:e02068. [CrossRef]
204. Javadzadeh SG, Asoodeh A. A novel textile dye degrading extracellular laccase from symbiotic bacterium of Bacillus sp. CF96 isolated from gut termite (Anacanthotermes). Int J Biol Macromol 2020;145:355-63. [CrossRef]
205. Carolin CF, Kumar PS, Joshiba GJ. Sustainable approach to decolourize methyl orange dye from aqueous solution using novel bacterial strain and its metabolites characterization. Clean Technol Environ Policy 2021;23:173-81. [CrossRef]
206. Yan J, Niu J, Chen D, Chen Y, Irbis C. Screening of trametes strains for efficient decolorization of malachite green at high temperatures and ionic concentrations. Int Biodeterior Biodegrad 2014;87:109-15. [CrossRef]
207. Price PB, Sowers T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci U S A 2004;101:4631-6. [CrossRef]
208. Fu Y, Viraraghavan T. Fungal decolorization of dye wastewaters:A review. Bioresour Technol 2001;79:251-62. [CrossRef]
209. Shedbalkar U, Dhanve R, Jadhav J. Biodegradation of triphenylmethane dye cotton blue by Penicillium ochrochloron MTCC 517. J Hazard Mater 2008;157:472-9. [CrossRef]
210. Telke AA, Joshi SM, Jadhav SU, Tamboli DP, Govindwar SP. Decolorization and detoxification of Congo red and textile industry effluent by an isolated bacterium Pseudomonas sp. SU-EBT. Biodegradation 2010;21:283-96. [CrossRef]
211. Ayed L, Mahdhi A, Cheref A, Bakhrouf A. Decolorization and degradation of azo dye Methyl Red by an isolated Sphingomonas paucimobilis:Biotoxicity and metabolites characterization. Desalination 2011;274:272-7. [CrossRef]
212. Zhang C, Diao H, Lu F, Bie X, Wang Y, Lu Z. Degradation of triphenylmethane dyes using a temperature and pH stable spore laccase from a novel strain of Bacillus vallismortis. Bioresour Technol 2012;126:80-6. [CrossRef]
213. Hussain S, Maqbool Z, Ali S, Yasmeen T, Imran M, Mahmood F, et al. Biodecolorization of reactive black-5 by a metal and salt tolerant bacterial strain Pseudomonas sp. RA20 isolated from Paharang drain effluents in Pakistan. Ecotoxicol Environ Saf 2013;98:331-8. [CrossRef]
214. Karunya A, Rose C, Nachiyar CV. Biodegradation of the textile dye Mordant Black 17 (Calcon) by Moraxella osloensis isolated from textile effluent-contaminated site. World J Microbiol Biotechnol 2014;30:915-24. [CrossRef]
215. Singh S, Chatterji S, Nandini PT, Prasad AS, Rao KV. Biodegradation of azo dye direct orange 16 by Micrococcus luteus strain SSN2. Int J Environ Sci Technol 2015;12:2161-8. [CrossRef]
216. El Bouraie M, El Din WS. Biodegradation of reactive Black 5 by Aeromonas hydrophila strain isolated from dye-contaminated textile wastewater. Sustain Environ Res 2016;26:209-16. [CrossRef]
217. Chukowry PK, Mudhoo A, Santchurn SJ. Bacillus algicola decolourises more than 95% of some textile azo dyes. Environ Chem Lett 2017;15:531-6. [CrossRef]
218. Meerbergen K, Willems KA, Dewil R, Van Impe J, Appels L, Lievens B. Isolation and screening of bacterial isolates from wastewater treatment plants to decolorize azo dyes. J Biosci Bioeng 2018;125:448-56. [CrossRef]
219. Ajaz M, Rehman A, Khan Z, Nisar MA, Hussain S. Degradation of azo dyes by Alcaligenes aquatilis 3c and its potential use in the wastewater treatment. AMB Expr 2019;9:64. [CrossRef]
220. Barathi S, Aruljothi KN, Karthik C, Padikasan IA. Optimization for enhanced ecofriendly decolorization and detoxification of reactive blue160 textile dye by Bacillus subtilis. Biotechnol Rep (Amst) 2020;28:e00522. [CrossRef]
221. Thangaraj S, Bankole PO, Sadasivam SK. Microbial degradation of azo dyes by textile effluent adapted, Enterobacter hormaechei under microaerophilic condition. Microbiol Res 2021;250:126805. [CrossRef]
222. Parshetti G, Kalme S, Saratale G, Govindwar S. Biodegradation of malachite green by Kocuria rosea MTCC 1532. Acta Chim Slov 2006;53:492-8.
223. Jirasripongpun K, Nasanit R, Niruntasook J, Chotikasatian B. Decolorization and degradation of C. I. reactive red 195 by Enterobacter sp. Sci Technol Asia 2007;12:6-11.
224. Gopinath KP, Sahib Ha, Muthukumar K, Velan M. Improved biodegradation of congored by using Bacillus sp. Bioresour Technol 2009;100:670-5. [CrossRef]
225. Jadhav JP, Phugare SS, Dhanve RS, Jadhav SB. Rapid biodegradation and decolorization of direct orange 39 (Orange TGLL) by an isolated bacterium Pseudomonas aeruginosa strain BCH. Biodegradation 2010;21:453-63. [CrossRef]
226. Unnikrishnan S, Khan MH, Ramalingam K. Dye-tolerant marine Acinetobacter baumannii-mediated biodegradation of reactive red. Water Sci Eng 2018;11:265-75. [CrossRef]
227. Mnif I, Maktouf S, Fendri R, Kriaa M, Ellouze S, Ghribi D. Improvement of methyl orange dye biotreatment by a novel isolated strain, Aeromonas veronii GRI, by SPB1 biosurfactant addition. Environ Sci Pollut Res 2016;23:1742-54. [CrossRef]
228. Chen G, An X, Feng L, Xia X, Zhang Q. Genome and transcriptome analysis of a newly isolated azo dye degrading thermophilic strain Anoxybacillus sp. Ecotoxicol Environ Saf 2020;203:111047. [CrossRef]
229. Kolekar YM, Pawar SP, Gawai KR, Lokhande PD, Shouche YS, Kodam KM. Decolorization and degradation of Disperse Blue 79 and Acid Orange 10, by Bacillus fusiformis KMK5 isolated from the textile dye contaminated soil. Bioresour Technol 2008;99:8999-9003. [CrossRef]
230. Kabeer FA, John N, Abdulla MH. Biodegradation of malachite green by a newly isolated Bacillus vietnamensis sp. MSB17 from continental slope of the Eastern Arabian Sea:Enzyme analysis, degradation pathway and toxicity studies. Bioremediat J 2019;23:334-42. [CrossRef]
231. Daâssi D, Mechichi T, Nasri M, Rodriguez-Couto S. Decolorization of the metal textile dye lanaset grey G by immobilized white-rot fungi. J Environ Manage 2013;129:324-32. [CrossRef]
232. Jadhav UU, Dawkar VV, Ghodake GS, Govindwar SP. Biodegradation of direct red 5B, a textile dye by newly isolated Comamonas sp. UVS. J Hazard Mater 2008;158:507-16. [CrossRef]
233. Dhanve RS, Shedbalkar UU, Jadhav JP. Biodegradation of diazo reactive dye navy blue HE2R (Reactive blue 172) by an isolated Exiguobacterium sp. RD3. Biotechnol Bioprocess Eng 2008;13:53-60. [CrossRef]
234. Kim SJ, Ishikawa K, Hirai M, Shoda M. Characteristics of a newly isolated fungus, Geotrichum candidum Dec 1, which decolorizes various dyes. J Ferment Bioeng 1995;79:601-7. [CrossRef]
235. Balamurugan B, Thirumarimurugan M, Kannadasan T. Anaerobic degradation of textile dye bath effluent using Halomonas sp. Bioresour Technol 2011;102:6365-9. [CrossRef]
236. Jafari N, Soudi MR, Kasra-Kermanshahi R. Biodecolorization of textile azo dyes by isolated yeast from activated sludge:Issatchenkia orientalis JKS6. Ann Microbiol 2014;64:475-82. [CrossRef]
237. Srinivasan S, Sadasivam SK. Exploring docking and aerobic-microaerophilic biodegradation of textile azo dye by bacterial systems. J Water Process Eng 2018;22:180-91. [CrossRef]
238. Bhattacharya A, Goyal N, Gupta A. Degradation of azo dye methyl red by alkaliphilic, halotolerant Nesterenkonia lacusekhoensis EMLA3:Application in alkaline and salt-rich dyeing effluent treatment. Extremophiles 2017;21:479-90. [CrossRef]
239. Prabhakar Y, Gupta A, Kaushik A. Enhanced decolorization of reactive violet dye 1 by halo-alkaliphilic Nesterenkonia strain:Process optimization, short acclimatization and reusability analysis in batch cycles. Process Saf Environ Prot 2019;131:116-26. [CrossRef]
240. Cheng H, Yuan M, Zeng Q, Zhou H, Zhan W, Chen H, et al. Efficient reduction of reactive black 5 and Cr(?) by a newly isolated bacterium of Ochrobactrum anthropi. J Hazard Mater 2021;406:124641. [CrossRef]
241. Feng C, Fang-Yan C, Yu-Bin T. Isolation, identification of a halotolerant Acid Red B Degrading strain and its decolorization performance. APCBEE Procedia 2014;9:131-9. [CrossRef]
242. Garg SK, Tripathi M, Singh SK, Tiwari JK. Biodecolorization of textile dye effluent by Pseudomonas putida SKG-1 (MTCC 10510) under the conditions optimized for monoazo dye orange II color removal in simulated minimal salt medium. Int Biodeterior Biodegrad 2012;74:24-35. [CrossRef]
243. Garg N, Garg A, Mukherji S. Eco-friendly decolorization and degradation of reactive yellow 145 textile dye by Pseudomonas aeruginosa and Thiosphaera pantotropha. J Environ Manage 2020;263:110383. [CrossRef]
244. Nor NM, Hadibarata T, Zubir MM, Lazim ZM, Adnan LA, Fulazzaky MA. Mechanism of triphenylmethane cresol red degradation by Trichoderma harzianum M06. Bioprocess Biosyst Eng 2015;38:2167-75. [CrossRef]
245. Saroj S, Dubey S, Agarwal P, Prasad R, Singh RP. Evaluation of the efficacy of a fungal consortium for degradation of azo dyes and simulated textile dye effluents. Sustain Water Resour Manage 2015;1:233-43. [CrossRef]
246. Chaudhari AU, Paul D, Dhotre D, Kodam KM. Effective biotransformation and detoxification of anthraquinone dye reactive blue 4 by using aerobic bacterial granules. Water Res 2017;122:603-13. [CrossRef]
247. Chen Y, Feng L, Li H, Wang Y, Chen G, Zhang Q. Biodegradation and detoxification of direct black G textile dye by a newly isolated thermophilic microflora. Bioresour Technol 2018;250:650-7. [CrossRef]
248. Amin S, Rastogi RP, Chaubey MG, Jain K, Divecha J, Desai C, et al. Degradation and toxicity analysis of a reactive textile diazo dye-direct red 81 by newly isolated Bacillus sp. DMS2. Front Microbiol 2020;11:576680. [CrossRef]
249. Rudakiya DM, Pawar KS. Optimization of culture condition for enhanced decolorization of reactive Orange 16 by Comamonas acidovorans MTCC 3364. Int J Curr Microbiol Appl Sci 2013;2:467-76.
250. Liu G, Zhou J, Meng X, Fu SQ, Wang J, Jin R, et al. Decolorization of azo dyes by marine Shewanella strains under saline conditions. Appl Microbiol Biotechnol 2013;97:4187-97. [CrossRef]
251. Xu F, Mou Z, Geng J, Zhang X, Li CZ. Azo dye decolorization by a halotolerant exoelectrogenic decolorizer isolated from marine sediment. Chemosphere 2016;158:30-6. [CrossRef]
252. Song L, Shao Y, Ning S, Tan L. Performance of a newly isolated salt-tolerant yeast strain Pichia occidentalis G1 for degrading and detoxifying azo dyes. Bioresour Technol 2017;233:21-9. [CrossRef]
253. Masarbo RS, Ismailsab M, Monisha TR, Nayak AS, Karegoudar TB. Enhanced decolorization of sulfonated azo dye methyl orange by single and mixed bacterial strains AK1, AK2 and VKY1. Bioremediat J 2018;22:136-46. [CrossRef]
254. Moharrery L, Otadi M, Miraly N, Zangeneh MM, Amiri R. Degradation of toluidine red, an oil soluble azo dye by Halomonas strain IP8 at alkaline condition. Chem Eng Commun 2019;206:61-8. [CrossRef]
255. Zhuang M, Sanganyado E, Zhang X, Xu L, Zhu J, Liu W, et al. Azo dye degrading bacteria tolerant to extreme conditions inhabit nearshore ecosystems:Optimization and degradation pathways. J Environ Manage 2020;261:110222. [CrossRef]
256. Sreedharan V, Saha P, Rao KV. Dye degradation potential of Acinetobacter baumannii strain VITVB against commercial azo dyes. Bioremediat J 2021;25:347-68. [CrossRef]
257. Sani RK, Azmi W, Banerjee UC. Comparison of static and shake culture in the decolorization of textile dyes and dye effluents by Phanerochaete chrysoporium. Folia Microbiol (Praha) 1998;43:85-8. [CrossRef]
258. Garg SK, Tripathi M. Microbial strategies for discoloration and detoxification of azo dyes from textile effluents. Res J Microbiol 2017;12:1-19. [CrossRef]
259. Husseiny SM. Biodegradation of the reactive and direct dyes using Egyptian isolates. J Appl Sci Res 2008;4:599-606.
260. Shah PD, Dave SR, Rao MS. Enzymatic degradation of textile dye reactive Orange 13 by newly isolated bacterial strain Alcaligenes faecalis PMS-1. Int Biodeterior Biodegrad 2012;69:41-50. [CrossRef]
261. Manai I, Miladi B, El Mselmi A, Smaali I, Ben Hassen A, Hamdi M, et al. Industrial textile effluent decolourization in stirred and static batch cultures of a new fungal strain Chaetomium globosum IMA1 KJ472923. J Environ Manage 2016;170:8-14. [CrossRef]
262. Popli S, Patel UD. Destruction of azo dyes by anaerobic-aerobic sequential biological treatment:A review. Int J Environ Sci Technol 2015;12:405-20. [CrossRef]
263. Nachiyar CV, Sunkar S, Kumar GN, Karunya A, Ananth P, Prakash P, et al. Biodegradation of acid blue 113 containing textile effluent by constructed aerobic bacterial consortia:Optimization and mechanism. J Bioremed Biodeg 2012;3:162. [CrossRef]
264. Ogugbue CJ, Sawidis T, Oranusi NA. Bioremoval of chemically different synthetic dyes by Aeromonas hydrophila in simulated wastewater containing dyeing auxiliaries. Ann Microbiol 2012;62:1141-53. [CrossRef]
265. Franca RD, Vieira A, Carvalho G, Oehmen A, Pinheiro HM, Crespo MT, et al. Oerskovia paurometabola can efficiently decolorize azo dye acid red 14 and remove its recalcitrant metabolite. Ecotoxicol Environ Saf 2020;191:110007. [CrossRef]
266. Sankaranarayanan A, Karthikeyan S, Markande A, Sharma A. Remazol reactive dye degrading Bacteria from freshwater fish of River Cauvery, Pallipalayam of Namakkal District, South India. Environ Syst Res 2021;10:29. [CrossRef]
267. Dawkar VV, Jadhav UU, Ghodake GS, Govindwar SP. Effect of inducers on the decolorization and biodegradation of textile azo dye Navy blue 2GL by Bacillus sp. VUS. Biodegradation 2009;20:777-87. [CrossRef]
268. Chen BY, Chen WM, Wu FL, Chen PK, Yen CY. Revealing azo-dye decolorization of indigenous Aeromonas hydrophila from fountain spring in Northeast Taiwan. J Chin Inst Chem Eng 2008;39:495-501. [CrossRef]