1. INTRODUCTION
Antibiotics have been the lifesaving drug for both humans and animals since the discovery of modern generation of penicillin. However, in the past several decades, the humankind has been facing a grievous problem of resistant microorganisms making various antibiotics ineffective on them [1,2]. As per the World Health Organization, about 170,000 people succumbed all over the world as a consequence of multidrug-resistant (MDR) tuberculosis infection [3,4]. Antimicrobial resistance (AMR) endures posing momentous communal health trouble in terms of fatality and financial loss [5]. Antibiotic resistance has been linked to ill use and overdose of medicines, continued use of same chemical formulations as drugs, dearth of new drug discovery by the pharmaceutical industry, all causing the development of superbugs [1,2,6].
In India age regulated mortality due to infectious disease was found to be 377/10,000 persons, which was highest in South Asia [7]. India has been named as “the AMR capital of the world” [8]. In India, over 70% isolates of gram-negative bacteria such as Acinetobacter baumannii, Escherichia coli, Klebsiella pneumoniae, and 50% of all the Pseudomonas aeruginosa species were found to be resistant to third-generation cephalosporins and fluoroquinolone. Among the Gram-positive bacteria, 10.5% of Enterococcus faecium were found to be resistant to vancomycin and 42.6% Staphylococcus aureus was reported as methicillin-resistant [9]. More than 700,000 people died per year while fighting with antimicrobial-resistant infections and it is predicted that by the end of 2050 additional 10 million people will die due to MDR microorganisms alone which is more than the people dying due to road accidents and cancer combined [10].
The acuteness and severity of antibiotic resistance are a disturbing condition for medical and scientific professionals. Researchers have to seek out for alternative medication or evolve novel drugs to encounter MDR microorganisms. An immediate urgency of therapeutic support is required for handling MDR microorganisms [11]. The WHO’s, 13 general program of work on effective traditional medicine from 2019 to 2023 stated that traditional and complementary medicines can make a significant contribution to the goal of achieving Universal Health Coverage to provide essential and emergency health services [12]. Traditional medicines based on plants are proving to be a good alternative for treatment as they are easily accessible with minimal or no side effects.
Plants are the treasure house of essential products for enduring human health in the primitive practice of medicine and present pharmaceutics worldwide [13]. In India, about 20,000 medicinal flora have been reported of which only 7000–7500 plants are in use by traditional practitioners for the medication of numerous infections and dietary supplements [14]. For the substitution of synthetic antimicrobial drugs, selection of plants as a resource for novel drugs is an alternative avenue open for the world. The plants having antimicrobial activity are associated with the occurrence of active compounds such as phenols, flavonoids, quinines, alkaloids, terpenoids, tannins, essential oil, glucosinolates, lignans, and antimicrobial peptides (AMPs) [15].
AMPs have gained tremendous consideration recently due to their vigorous killing action on microbes. AMPs have been labeled as evolutionary earliest weapons’ to fight microbial infection. AMPs perform a significant activity in innate immunity of prokaryotic (microorganisms) and eukaryotic (insects, plant, and animals) system by regulating the first line of defense against pathogens [16]. AMPs are positively charged molecules containing disulfide bonds. These peptides hinder the proliferation of microorganisms by creating ion channels in microbial membrane, after a while cause the microbial cell to die [11]. AMPs have multifarious killing mechanism on microbes that includes obstruction in the DNA replication, transcription, translation and leading to cell membrane permeabilization which ultimately leads to death of the microbes.
Plants are one of the major sources of AMPs. Plant AMP (PAMPs) have significant accountability in the host defense mechanism, similar to human AMPs [11,17]. PAMPs are short cysteine-rich cationic peptides (10–50 amino acids) with antimicrobial activities that possess molecular weight of around 2–9 kDa. These peptides primarily constitute of hydrophobic amino acid with helical structure [18]. PAMPs can be a primary element for the development of new antibiotics, which may be competent against MDR infections, presently problematic to cure [19]. Grounded on the above facts, the present research was anticipated to isolate, purify, and characterize MoCh I AMP from Momordica charantia seeds.
2. MATERIALS AND METHODS
2.1. Collection of M. charantia Seeds
Fruits of M. charantia commonly called as Bitter Gourd were collected from vegetable markets. Fruits were allowed to ripen and then seeds were extracted. After air-drying, seeds were ground and kept in deep freezer until use. Seeds of M. charantia were verified from the CSIR - National Institute of Science Communication and Information Resources, New Delhi.
2.2. Collection of Microorganisms
The microbes used in the present work were Bacillus subtilis microbial type culture collection (MTCC 1789), Bacillus licheniformis (MTCC 1483), Candida albicans (MTCC 183), E. coli (MTCC 118), K. pneumoniae (MTCC 109), and S. aureus (MTCC 96). The microbial strains were acquired from the MTCC, institute of microbial technology, Chandigarh.
2.3. Isolation of MoCh I Peptide
For crude extract, 10 g seed powder was added to 150 mL phosphate buffer saline (PBS, pH 7.2) buffer. Mixture was refrigerated at −20°C succeeded by defrosting while maintaining at 4°C. Freezing-thawing approach was driven repeatedly for 3–4 days. The concentrate was spun at 10,000 rpm at 4°C for 30 min. The supernatant was strained using Whatman® filter paper 1 and the filtrate was saturated with 75% ammonium sulfate while maintaining at 4°C accompanied by centrifugation at 10,000 rpm for 30 min [20]. After centrifugation, precipitated protein pellet was mixed in PBS buffer and supernatant was kept in deep freezer until use.
2.4. Protein Quantitation
The protein content present in protein pellet and supernatant collected from above step was detected by Bradford method using bovine serum albumin as standard [21].
2.5. Antimicrobial Assay
The antimicrobial activity was estimated through agar well diffusion method [22]. For antibacterial and antifungal activity, nutrient agar and yeast peptone dextrose media were used, respectively. A lawn was set with activated culture using sterile cotton swabs. Well was created and dissolved protein pellet and supernatant were added. Petri plates were incubated at 27°C and 37°C for fungal and bacterial culture respectively for 24 h. For positive control samples, Tertracyclin disc (30 μg) against K. pneumonia, Fluconazole (70 μg) against C. albicans, Chloramphenicol (50 μg) against B. subtilis and B. licheniformis, and Ampicillin (60 μg) against E. coli and S. aureus were used. For negative control, PBS buffer was used. The zone of inhibition was measured in millimeters (mm).
2.6. Purification of MoCh I Peptide
The protein pellets having antimicrobial activity were dialyzed using Tube-O-Dialyzer™ (G-Biosciences) of MWCO 1 kDa, followed by Amicon® Ultra filter (Merck) of MWCO 10 kDa. After centrifugation, the protein <10 kDa was collected and lyophilized. The lyophilized sample was dissolved in milliQ water and stored at −20°C. Antimicrobial activity of lyophilized MoCh I peptide was carried out.
2.7. Biological Activity of MoCh I Peptide
2.7.1. Minimum inhibitory concentration (MIC)
Based on the antimicrobial activity recorded earlier, the MIC of MoCh I peptide was determined on selected gram-positive bacteria (B. licheniformis, B. subtilis, and S. aureus). The 0.5 Mcfarland standard solution was used to optimize the turbidity of bacterial culture. The 0.5 McFarland standard had absorbance ranging between 0.08 and 0.2 at 625 nm. The bacterial culture after turbidity adjustment was added to each well. For negative control, the bacterial culture and bacterial culture along with antibiotic was used as positive control. In test sample, different concentration of purified MoCh I peptide was added to bacterial culture. Plate was incubated for 12–16 hours at 37°C. After incubation, optical density of the 96-well microtiter tray was recorded at 600 nm using ELISA reader [23].
2.7.2. Erythrocyte hemolysis assay
EDTA coated vials were used for collecting Blood and it was then centrifuged at 1500 rpm for 5 min. Erythrocytes were washed twice with PBS buffer. The 10% erythrocytes suspension in PBS was incubated with various concentrations of purified peptides. The mixture of erythrocytes and purified peptides were placed in a 5% CO2 incubator at 37°C for 24 h. After incubation, the sample was centrifuged at 1500 rpm for 5 min, supernatant was shifted to ELISA plate and optical density was measured at 405 nm in ELISA reader. A PBS buffer and 1% Triton X-100 (G-Biosciences) were used as negative and positive control, respectively [24].
2.8. Characterization of MoCh I Peptide
2.8.1. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (tricine- SDS-PAGE)
The Tricine SDS-PAGE was executed by Schagger approach using 16% separating/resolving gel and 5% stacking gel [25]. The lyophilized protein sample was mixed with the sample buffer followed by heat denaturation at 95°C for 2 min and then loaded into the wells. The gel was run at 120 V and stained with Coomassie Brilliant Blue staining solution. The molecular weight of MoCh I peptide was determined using standard protein ranging between 1.06 kDa and 26.6 kDa (Sigma).
2.8.2. Matrix assisted laser desorption/ionization-time of flight/time of flight mass spectrometry (MALDI-TOF/TOF MS/MS)
The gel plugs were taken from the Tricine SDS-PAGE gel for digestion. Peptide was extracted from the gel plugs using standard method [26]. A 1:1 ratio of peptides obtained above was obtained with α-Cyano-4-hydroxycinnamic acid matrix. The resulting mixture was dotted upon the MALDI plate. The sample was air dried and examined on the MALDI TOF/TOF Ultraflex III instrument (Bruker Daltonics, Germany). Extrinsic adjustment was carried out with standard peptide (PEPMIX Mixture) provided by Bruker, with masses varying from 1046 to 3147 Da. The evaluation was carried out using Flex analysis software in reflectron ion mode. The Mascot search in “Firmicutes” database was used for identification of the protein from the masses obtained in protein mass fingerprinting (PMF).
2.8.3. Structure determination of MoCh I peptide
The secondary structure of MoCh I peptide was determined by circular dichorism (CD) spectra at 25°C on a Jasco CD spectropolarimeter equipped with peltier based computer driven temperature control system and examined by Jasco software. The MoCh I peptide was scanned between 198 and 250 nm wavelength having 0.2 cm cell path length [24].
2.8.4. In silico identification of MoCh I peptide
Protein – Protein BLAST (BLASTp) was performed using the peptide sequence obtained from De-novo sequencing (Peptide Mass Fingerprinting) [27]. Conserved domain database (CDD) was used for the identification of conserved domains for the function prediction of MoCh I peptide [28]. InterProScan provides the functional evaluation of proteins by arranging them into families and forecast domains and important sites [29]. Simple modular architecture research tool (SMART) tool was used to determine the homologous super-family, Pfam domains, signal peptides and internal peptides of MoCh I peptide [30]. Protein Homology/analogy Recognition Engine V 2.0 (PHYRE2) was used for the prediction of 3-D structure of MoCh I peptide. The amino acid sequence of peptide using intensive modeling model was searched for 3-D structure prediction [31]. iCn3D Viewer was used to visualize the 3D structure of protein. The viewer showed the alignment of MoCh I peptide sequence to a homologous structure [32].
2.9. Statistical Analysis
All the experiments were executed in triplets. All graphs and statistical analysis were executed in MS Excel. The data were expressed as Mean ± SD. Student t-test was accomplished to find the significance of data (**P < 0.05; ***P < 0.01).
3. RESULTS AND DISCUSSION
3.1. Protein Concentration
The protein concentration of 75% saturated protein pellet and supernatant was determined by Bradford method. In seed extract of M. charantia, protein concentration in pellet and supernatant was found to be 726.0 ± 33.94 μg/mL and 273.5 ± 33.23 μg/mL, respectively.
3.2. Antimicrobial Activity of 75% Saturated Protein Pellet
The protein pellet and supernatant obtained after 75% ammonium sulfate saturation was screened for antimicrobial activity. The protein pellet had 5.05 ± 0.07 mm, 3.60 ± 0.14 mm, and 2.55 ± 0.07 mm zone of inhibition against B. subtilis, B. licheniformis, and S. aureus, respectively. The dialyzed protein pellet (MWCO 1 kDa) had antimicrobial activity with zone of inhibition 3.25 ± 0.35 mm, 3.50 ± 0.70 mm, and 4.25 ± 0.3535 mm against B. subtilis, B. licheniformis, and S. aureus, respectively [Table 1 and Figure 1]. The 75% saturated protein pellet did not show any antimicrobial activity against selected Gram-negative bacteria (E. coli and K. pneumoniae) and fungi (C. albicans). The undialyzed and dialyzed supernatant obtained after saturation does not have antimicrobial activity against any of the selected microorganisms.
Table 1: Antimicrobial activity of 75% saturated protein pellet against selected microorganisms.
| S. No. | 75% ammonium sulfate saturated protein pellet | Zone of inhibition in millimeters (mm) (mean±SD) | |||||
|---|---|---|---|---|---|---|---|
| Bacillus subtilis | Bacillus licheniformis | Staphylococcus aureus | Escherichia coli | Klebsiella pneumoniae | Candida albicans | ||
| 1. | Undialyzed | 5.05±0.07 | 3.60±0.14 | 2.55±0.07 | 0 | 0 | 0 |
| 2. | Dialyzed | 3.25±0.35 | 3.50±0.70 | 4.25±0.35 | 0 | 0 | 0 |
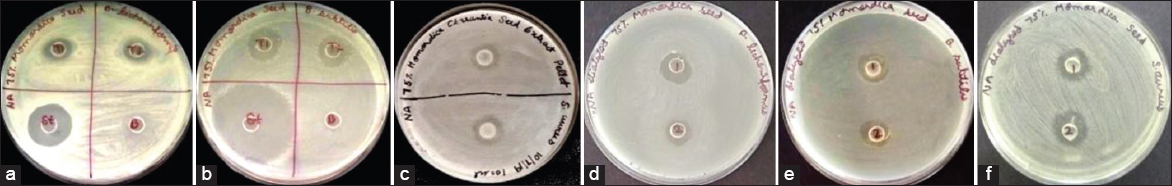 | Figure 1: Antimicrobial activity against B. licheniformis, B. subtilis and S. aureus of M. charantia undialyzed protein pellet (a-c); dialyzed protein pellet (d-f). [Click here to view] |
The action of AMPs mainly is their ability to collapse the membrane by interacting with lipid molecules on bacterial surface and/or interacting with other targets in the cytoplasm and that too depends on many factors, such as charge, size, and folding pattern of peptides leading to channel (pore) formation [33-36]. The antimicrobial activity depends on the physical and chemical properties of the peptide and also the phospholipids present on the bacterial membrane [37]. The AMPs also act by aggregating on the membrane, leading to the formation of “bends” in the membrane, causing the pore formation [38,39]. The disruption of membrane leads to pore formation, and this eventually causes leakage of ions and metabolites and disrupts the respiratory mechanism and synthesis of cell wall, finally leading to collapse of membrane [34,40].
The microorganisms sometimes may survive for prolonged periods even after the membrane have been permeabilized, signifying that mechanisms other than those involving the membrane are also responsible for cell death. These non-membranolytic mechanisms include, binding of PAMPs to other factors such as replicating enzymes, transcription factors, translation initiation factors, cofactors, and receptors (CCR2, CCR6, and FPLR1), lead to cessation of DNA synthesis, transcription and translation process and ultimately leads to cell death [34,41]. They can easily penetrate into the cell through membrane permeabilization causing the death of microorganisms. Due to the diverse activity of AMPs on microbes, the likelihood of the microbes becoming MDRs are less.
The negative result recorded against selected fungi (C. albicans) and Gram-negative bacteria (E. coli) indicate that these proteins/peptides might not have been to interact with the membrane proteins or lipids of the mentioned microorganisms. It has already been shown that glucosylceramides (GlcCer) present in the membrane of fungi, binds with the peptides causing oxidative damage to cell and cause induction of reactive oxygen species and nitric oxide production [42,43]. In the present study, probably non-interaction of the peptides with GlcCer present on the microorganism (C. albicans) caused non-permeabilization of the membrane thereby showing no antimicrobial activity [44]. In case of gram-negative bacteria, peptides interact with the lipopolysacharides and proteins like omptins present on the membrane. The omptins, which are aspartate and serine proteases bound on the outer membrane or secreted by these Gram-negative bacteria. The active AMPs can be converted into inactive fragments by bacterial proteases that lead to host bacteria becoming resistant toward AMPs [45-47].
3.3. Purification of AMPs
The 75% saturated protein pellet of M. charantia was dialyzed through 1 kDa MWCO dialysis membrane followed by Amicon Ultra centrifugal filtration (10 kDa MWCO). The double dialyzed protein pellet was lyophilized and dissolved in MilliQ water. Before lyophilization, the protein concentration was found to be 0.29 ± 0.01 mg/mL, and after lyophilization it was found to be 8.70 ± 0.35 mg/mL. The lyophilized protein pellet was analyzed on 16% Tricine SDS PAGE and a single band of approximately 3.70 kDa was obtained on the gel [Figure 2].
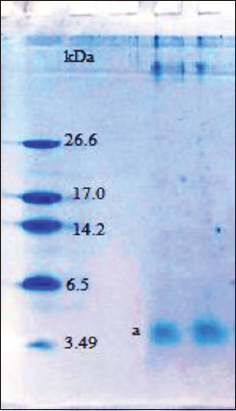 | Figure 2: Tricine SDS-PAGE of purified MoCh I peptide showing band of 3.7 kDa. [Click here to view] |
3.4. Biological Testing of Purified MoCh I Peptide
3.4.1.MIC
The MIC was carried out using ELISA reader. The overnight grown broth culture was inoculated with different concentrations of purified peptide and incubated at 37°C overnight. The optical density of treated cultures was recorded at 600 nm using ELISA reader. The MIC was found to be 65.2 μg/mL against B. subtilis whereas, MIC value against B. licheniformis and S. aureus was recorded as 87.0 mg/mL [Figure 3].
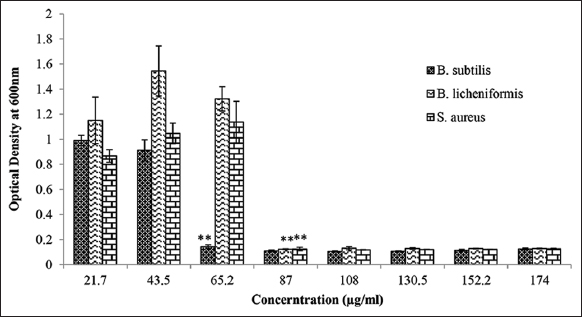 | Figure 3: MIC of purified MoCh I peptide against B. subtilis, B. licheniformis, and S. aureus. The data were found to be statistically significant (**P < 0.05). [Click here to view] |
3.4.2. Erythrocyte hemolysis assay
Different concentrations of purified peptide were added to 10% erythrocyte suspension and the incubation was carried out at 37°C in 5% CO2 incubator for 24 h. After 24 h, optical density at 405 nm was measured. The red blood cells were treated with 1% Triton X-100 resulted in complete lysis of RBCs. Hemolysis of RBCs was not observed with 21.7 μg/mL–130.5 μg/mL concentration of purified peptide. Whereas, 51.24% and 61.09% lysis of RBCs was observed with 152.2 μg/mL and 174 μg/mL concentration of purified peptide, respectively [Figure 4]. The previous reports suggested that if hemolysis did not occur at MIC value of the peptide, then it could be used as a drug in therapeutic treatments [48]. In our study, the concentration of peptide showing around 50% hemolysis was much higher than the MIC value of peptide indicating no hemolysis of RBCs at MIC value. From the above results, it could be inferred that purified peptide has antimicrobial and non-hemolytic properties and is safe to be used as drug. This single purified peptide from the seed extract of M. charantia was named as MoCh I peptide.
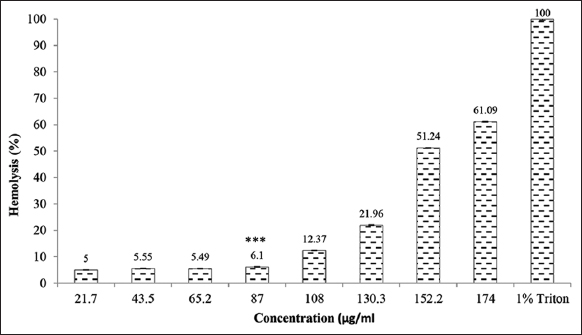 | Figure 4: Hemolysis using purified MoCh I peptide. The data were found to be statistically significant (***P < 0.01). [Click here to view] |
3.5. Characterization of MoCh I Peptide
3.5.1. Sequence of MoCh I peptide using MALDI-TOFF MS/MS
The protein sample having MoCh I peptide was separated on Tricine SDS-PAGE. The band having MoCh I peptide was subjected to in-gel digestion after extraction from gel. The MoCh I peptide was fragmented by trypsination and then resolved by MALDI-TOFF MS and MS/MS [Figures 5 and 6]. In the present study, Mascot search engine and SwissProt database was used for PMF. The list of protein with protein score was created by Mascot search engine. For MoCh I peptide, top score was found to be 117 for trypsin inhibitor 1 (MCoTI-I/ITR1_MOMCO) isolated from Momordica cochinchinensis. The MS/MS spectrum of MoCh I peptide sequence was found to have similarity with trypsin inhibitors of M. cochinchinensis.
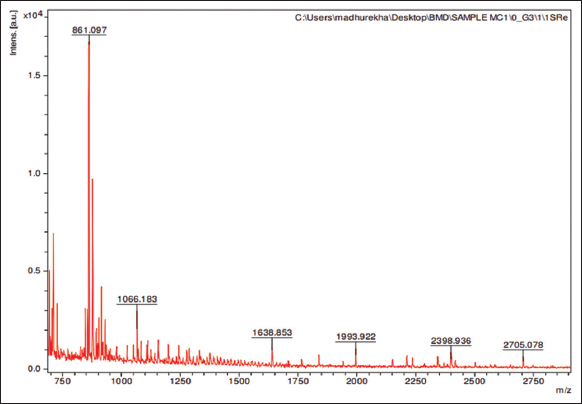 | Figure 5: MALDI-TOFF MS spectrum of MoCh I peptide. [Click here to view] |
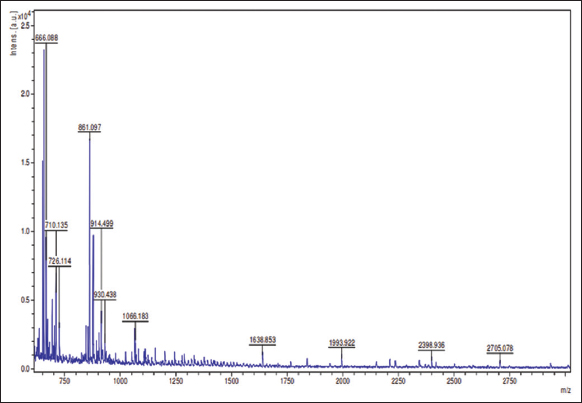 | Figure 6: MALDI-TOFF MS/MS spectrum of MoCh I peptide. [Click here to view] |
The MoCh I was found to be have sequence similarity with Trypsin inhibitor 1 of M. cochinchinensis. The MoCh I peptide was recorded to have 100% sequence coverage with MCoTI-I/ITR1_MOMCO trypsin inhibitor protein. The de novo sequencing revealed 34 amino acid (SGSDGGVCPK ILQRCRRDSD CPGACICRGN GYCG) sequence of MoCh I peptide, which was similar to MCoTI-I/ITR1_MOMCO.
The Mascot search indicated high sequence homology and top score for trypsin inhibitor 1 (MCoTI-I/ITR1_MOMCO) isolated from M. cochinchinensis, which is a Knottin type AMP. The conserved disulfide-bonding pattern was depicted as CysI-CysIV, CysII-CysV, and CysIII-CysVI which is normally observed in Knottin type peptide. Therefore, the predicted disulfide bonding pattern for MoCh I peptide should be same as MCoTI-I/ITR1_MOMCO trypsin inhibitor protein. The MCoTI-I/ITR1_MOMCO was reported under squash type serine protease inhibitor family (Protease inhibitor I7) has an important role in plant defense mechanism [49]. The MoCh I Trypsin inhibitor peptide comprises 34 amino acid residues. The occurrence of “cyclic cystine knot” structurally describes MoCh I protein as a Knottin [50]. The reactive bond is present between 10th and 11th amino acid residue. The cyclopeptide crosslink was present between 1st serine and 34th glycine amino acid residue. The helix and strand was present between 18th–20th and 31st–34th amino acid residues, respectively. The previous reports also suggest that squash trypsin inhibitor subfamily is frequently considered under Cyclotide subfamily [51,52].
3.6. In silico Identification of MoCh I Peptide
3.6.1. Protein basic local alignment search tool (BLASTp)
The significant sequence retrieved after BLAST corresponded to squash trypsin inhibitor MCoTI-II/ITR2_MOMCO (M. cochinchinensis) having 94.12% similarity with 100% sequence coverage. Another significant sequence was MCoTI-I/ITR1_MOMCO which showed 100% sequence similarity with 88% sequence coverage. The trypsin inhibitor I and III (MCTI-I and MCTI-III) from the M. charantia showed 79% sequence similarity and 66.67–62.96% sequence coverage. The putative conserved domain having inhibitory residue Lysine (K) at 10th position belonging to Plant Trypsin inhibitor (Plant TI) super family was retrieved. The sequence alignment of MoCh I and significant database sequence showed that there was substitution of amino acids at 13th and 14th position in which Lysine (basic) was substituted with Glutamine (neutral) and Arginine (strongly basic), respectively. Since the physicochemical property remains the same therefore, we do not expect any change in the structure or folding pattern of MoCh I peptide. Sequences of the species showing significant similarity to the query sequence (MoCh I) were selected from the BLASTp output and were further subjected to Phylogenetic alignment using Neighbor joining. In the phylogenetic distance tree, sequence similarity was observed between MoCh I trypsin inhibitor and trypsin inhibitors of M. cochinchinensis [Figure 7].
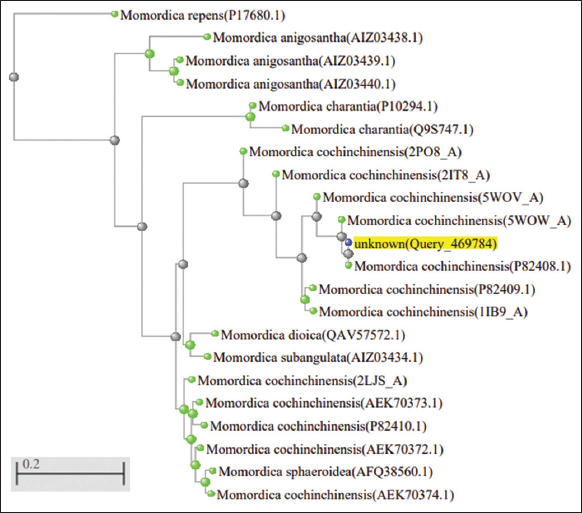 | Figure 7: Phylogenetic tree for MoCh I peptide sequence. [Click here to view] |
3.6.2. Conserved domains
The CDD, InterProScan, and SMART tool revealed that MoCh I peptide consist of conserved domain which belongs to Plant TI domain. The InterProScan also indicated MoCh I peptide as homologous to Proteinase amylase inhibitor super-family. Gene ontology predicted the molecular function of MoCh I peptide as Serine type endopeptidase inhibitor activity. In addition to the above information, SMART tool also predicted that MoCh I peptide showed homology with Squash Trypsin inhibitor domain present in the protein of M. cochinchinensis (MCoTI-I/ITR1_MOMCO) species and other species of cucurbitaceae family [53,54].
3.7. Structure of MoCh I Peptide
The structure of MoCh I pepetide was predicted by circular dichroism (CD). In CD spectra, a small positive peak was centered at 217 nm and large negative peak was observed at 198 nm [Figure 8]. The structure of MoCh I peptide was also predicted in-silico using PHYRE2 tool [Figure 9]. The secondary structure prediction showed the presence of 24% α-helix, 12% β-sheet, and 38% disordered regions in the structure of MoCh I peptide. The putative structure of MoCh I peptide was also visualized using iCn3D viewer. The MoCh I peptide structure was homologous to structure of MCoTI-II/ITR2_MOMCO trypsin inhibitor isolated from M. cochinchinensis (PDB ID: 1HA9) [Figure 10]. The structure showed sequence homology between the MCoTI-II/ITR2_MOMCO and MoCh I peptide. The three disulfide bonds between Cys8-Cys25, Cys15- Cys27, and Cys21-Cys33 and two sheets (secondary structure) between Ile26-Gly29 and Gly31-Gly34 were present in the structure. In sequences and annotations, red color sequences showed homology, blue color showed substitution of amino acid with such as amino acid, presence of three disulfide bonds and single inhibitory site at 10th amino acid (Lysine). The 27 residues from 8th to 34th amino acid in MoCh I peptide showed the presence of Plant TI family conserved domain.
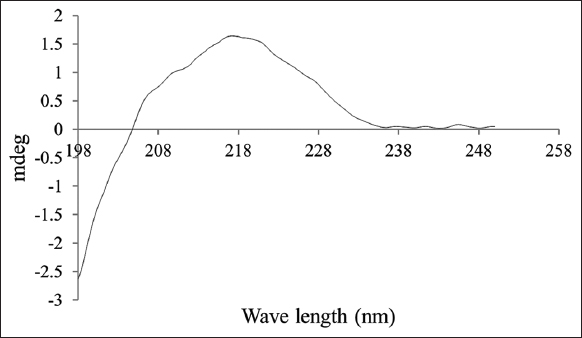 | Figure 8: CD spectra of MoCh I peptide. [Click here to view] |
 | Figure 9: Putative 3-D structure of MoCh I peptide using PHYRE2 tool. [Click here to view] |
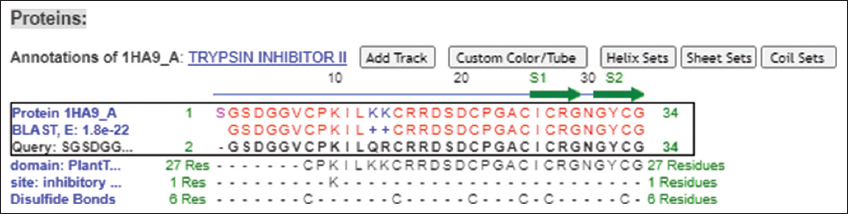 | Figure 10: Sequences and annotations of putative MoCh I peptide structure using iCn3D viewer. [Click here to view] |
The structure of MoCh I peptide as shown by CD was a small positive peak at 217 nm and large negative peak at 198 nm. This indicated the presence of β-sheets and turn in the structure of MoCh I peptide. It was in accordance with previously reported structures. According to this report, negative peaks between 190 and 200 nm and positive transition peaks between 200 and 205 nm indicated the presence of β-sheets and β-turn in the structure of the protein [55]. The absence of negative peak in the region of 200–240 nm pointed towards the absence of α-helix. This could be due the cyclic nature of MoCh I peptide as the cyclic peptides are unstable and unfold very fast. Due to structural similarity, MoCh I trypsin inhibitor might follow the same mechanism of action as described by Squash Trypsin inhibitor super-family members which bind the lipid membrane of microorganism and leads to membrane permeabilization [56]. However, MoCh I trypsin inhibitor peptide have only 79% sequence similarity with already reported trypsin inhibitor (MCTI-A) of M. charantia [57,58]. This might be due to different variety of M. charantia, climatic conditions of geographical area and stress conditions (salt, microbial, heavy metals, etc.) during plant growth. It has been reported that stress (salt, temperature, heavy metal, etc.) or variation in climatic conditions leads to change in antimicrobial protein concentration [59]. In India, two varieties of M. charantia namely var. charantia and var. muricata are grown whereas five different cultivars (Abashi-goya, Naga-goya, Shiro-goya, Nikko, and Peacock) of M. charantia are grown in Japan and China [56,60]. The use of different cultivars of M. charantia leads to production of different amino acids composition ultimately forming different proteins/peptides [56]. Therefore, the reason behind the low sequence similarly between MoCh I and MCTI-A peptide might be due to the different cultivars of M. charantia.
4. CONCLUSION
It has been observed that, the human pathogenic bacteria have evolved to develop resistance against AMPs during the course of evolution. However, the same is not applicable for the PAMPs. Plants vary from animals in their protein modification system and therefore are less amenable to human pathogens. Plants can be an appealing choice as they themselves are not under attack by human pathogens; hence, it is unlikely that these pathogens will get exposed to PAMPs and/or modify themselves to become resistant against PAMPs. The isolated novel PAMP (MoCh I) having molecular weight of 3.7 kDa from M. charantia is a prospective candidate to be developed as drug for the cure of microbial infections. Major feature of our novel peptide is its small size, which might give it a stronger penetrating power in the membrane of microorganism making it more potent candidate for drug development. However, further studies on stability and drug delivery system need to be carried out to prove its efficacy against various pathogenic and MDR microorganisms.
5. AUTHORS’ CONTRIBUTIONS
All authors contributed to the design and conception of this study and participated in the writing and editing of the manuscript. S, RB, and KB executed and analyzed the wet-bench results; RK and SS help in analyzing in silico results. All authors read and approved the final manuscript.
6. FUNDING
There is no funding to report.
7. CONFLICT OF INTEREST
All the authors also declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
8. ETHICAL APPROVAL
The present study does not contain any studies with human participants or animals performed by any of the authors.
9. DATA AVAILABILITY
Data generated and analyzed during this study are included in the published article.
10. ACKNOWLEDGMENT
We acknowledge the DAV Management Committee, India and DAV Central Instrumentation Laboratory funded by DST-FIST, India, for the infrastructure support.
11. CONSENT OF PUBLICATION
All the authors have gone through the manuscript and have consented to proceed with publication in this journal.
REFERENCES
1. Centers for Disease Control and Prevention. Office of Infectious Disease. Antibiotic Resistance Threats in the United States. United States:Centers for Disease Control and Prevention;2013.
2. Read AF, Woods RJ. Antibiotic resistance management. Evol Med Public Health 2014;2014:147. [CrossRef]
3. World Health Organization. Jaipur Declaration on Antimicrobial Resistance. Geneva:World Health Organization;2011.
4. Gross M. Antibiotics in crisis. Curr Biol 2013;23:R1063-5. [CrossRef]
5. Taneja N, Sharma M. Antimicrobial resistance in the environment:The Indian scenario. Indian J Med Res 2019;149:119-28. [CrossRef]
6. Ventola CL. The antibiotic resistance crisis:Part 1:Causes and threats. P T 2015;40:277-83.
7. World Health Organization. Research for International Tobacco Control. WHO Report on the Global Tobacco Epidemic, 2008:The MPOWER Package. Geneva:World Health Organization;2008.
8. Chaudhry D, Tomar P. Antimicrobial resistance:The next BIG pandemic. Int J Community Med Public Health 2017;4:2632-6. [CrossRef]
9. Gandra S, Joshi J, Trett A, Lamkang AS, Laxminarayan R. Scoping Report on Antimicrobial Resistance in India. United States:Center for Disease Dynamics, Economics and Policy;2017.
10. O'Neill J. Tackling Drug-Resistant Infections Globally:Final Report and Recommendations. London:Wellcome Trust and HM Government;2016.
11. Chandra H, Bishnoi P, Yadav A, Patni B, Mishra AP, Nautiyal AR. Antimicrobial resistance and the alternative resources with special emphasis on plant-based antimicrobials-a review. Plants (Basel) 2017;6:16. [CrossRef]
12. World Health Organization. Global Status Report on Alcohol and Health 2018. Geneva:World Health Organization;2019.
13. Singh AR, Bajaj VK, Sekhawat PS, Singh K. Phytochemical estimation and antimicrobial activity of aqueous and methanolic extract of Ocimum sanctum L. J Nat Prod Plant Resour 2013;3:51-8.
14. Pandey MM, Rastogi S, Rawat AK. Indian herbal drug for general healthcare:An overview. Int J Altern Med 2008;6:3. [CrossRef]
15. Golla K, Vutukuru SS, Rani JU, Meghanath P, Pasha C. Screening of small peptides from various germinating seeds having antimicrobial activity. J Pharm Biomed Anal 2006;11:2278-3008.
16. Yili A, Maksimov V, Ma QL, Gao YH, Veshkurova O, Salikhov S, et al. Antimicrobial peptides from the plants. J Pharm Pharmacol 2014;2:627-41. [CrossRef]
17. Company N, Nadal A, Ruiz C, Pla M. Production of phytotoxic cationic α-helical antimicrobial peptides in plant cells using inducible promoters. PLoS One 2014;9:e109990. [CrossRef]
18. Nawrot R, Barylski J, Nowicki G, Broniarczyk J, Buchwald W, Go?dzicka-Józefiak A. Plant antimicrobial peptides. Folia Microbiol (Praha) 2014;59:181-96. [CrossRef]
19. Samriti, Biswas R, Biswas K. Plant antimicrobial peptides:a novel approach against drug resistant microorganisms. Int J Pharm Sci Research. 2018;9:1-5.
20. Rehman S, Khanum A. Isolation and characterization of peptide (s) from Pisum sativumhaving antimicrobial activity against various bacteria. Pak J Bot 2011;43:2971-8.
21. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248-54. [CrossRef]
22. Reeves DS, Hawkey PM, Lewis DA. Antibiotic assays. Medical Bacteriology:A Practical Approach. United Kingdom:Oxford University Press;?2005.
23. Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother 2002:49:1049. [CrossRef]
24. Baindara P, Chaudhry V, Mittal G, Liao LM, Matos CO, Khatri N, et al. Characterization of the antimicrobial peptide penisin, a class Ia novel lantibiotic from Paenibacillus sp. strain A3. Antimicrob Agents Chemother 2016;60:580-91. [CrossRef]
25. Schägger H. Tricine-SDS-PAGE. Nat Protoc 2006;1:16-22. [CrossRef]
26. Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 2006;1:2856-60. [CrossRef]
27. National Center for Biotechnology Information. U.S. National Library of Medicine. United States:National Center for Biotechnology Information. Available from:https://blast.ncbi.nlm.nih.gov/Blast.cgi
28. Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD:A database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res 2002;30:281-3. [CrossRef]
29. Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, et al. The InterPro protein families and domains database:20 years on. Nucleic Acids Res 2021;49:D344-54. [CrossRef]
30. Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool:Identification of signaling domains. Proc Natl Acad Sci U S A 1998;95:5857-64. [CrossRef]
31. Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015;10:845-58. [CrossRef]
32. Wang J, Youkharibache P, Zhang D, Lanczycki CJ, Geer RC, Madej T, et al. iCn3D, a Web-based 3D viewer for sharing 1D/2D/3D representations of biomolecular structures. Bioinformatics 2020;36:131-5. [CrossRef]
33. Epand RM, Vogel HJ. Diversity of antimicrobial peptides and their mechanisms of action. Biochim Biophys Acta 1999;1462:11-28. [CrossRef]
34. Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev 2003;55:27-55. [CrossRef]
35. Gazit E, Boman A, Boman HG, Shai Y. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry 1995;34:11479-88. [CrossRef]
36. Matsuzaki K, Murase O, Fujii N, Miyajima K. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 1996;35:11361-8. [CrossRef]
37. Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense?Magainins and tachyplesins as archetypes. Biochim Biophys Acta 1999;1462:1-10. [CrossRef]
38. Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Membrane pores induced by magainin. Biochemistry 1996;35:13723-8. [CrossRef]
39. Silva PM, Gonçalves S, Santos NC. Defensins:Antifungal lessons from eukaryotes. Front Microbiol 2014;5:97. [CrossRef]
40. Barbosa Pelegrini P, Del Sarto RP, Silva ON, Franco OL, Grossi-de-Sa MF. Antibacterial peptides from plants:What they are and how they probably work. Biochem Res Int 2011;2011:250349. [CrossRef]
41. Sechet E, Telford E, Bonamy C, Sansonetti PJ, Sperandio B. Natural molecules induce and synergize to boost expression of the human antimicrobial peptide b-defensin-3. Proc Natl Acad Sci U S A 2018;115:E9869-78. [CrossRef]
42. Aerts AM, François IE, Meert EM, Li QT, Cammue BP, Thevissen K. The antifungal activity of RsAFP2, a plant defensin from Raphanus sativus, involves the induction of reactive oxygen species in Candida albicans. J Mol Microbiol Biotechnol 2007;13:243-7. [CrossRef]
43. Mello EO, Ribeiro SF, Carvalho AO, Santos IS, Da Cunha M, Santa-Catarina C, et al. Antifungal activity of PvD1 defensin involves plasma membrane permeabilization, inhibition of medium acidification, and induction of ROS in fungi cells. Curr Microbiol 2011;62:1209-17. [CrossRef]
44. Kiba A, Saitoh H, Nishihara M, Omiya K, Yamamura S. C-terminal domain of a hevein-like protein from Wasabia japonica has potent antimicrobial activity. Plant Cell Physiol 2003;44:296-303. [CrossRef]
45. Samriti, Biswas R, Biswas K. Isolation and characterization of antimicrobial peptides from Datura inoxia leaves having antimicrobial activity against selected bacteria. World J Pharm Res 2018;7:783-94.
46. Samriti, Biswas R, Biswas K. Antibacterial activity of antimicrobial peptide extracted from Trianthema portulacastrum Leaves. Pharma Innov J 2019;8:81-6.
47. Samriti, Biswas R, Biswas K. Antimicrobial proteins/peptides, isolated from two cultivars of Bougainvillea. Int J Pharm Sci Res 2021;12:604-14.
48. King A, Chakrabarty S, Zhang W, Zeng X, Ohman DE, Wood LF, et al. High antimicrobial effectiveness with low hemolytic and cytotoxic activity for PEG/quaternary copolyoxetanes. Biomacromolecules 2014;15:456-67. [CrossRef]
49. Hernandez JF, Gagnon J, Chiche L, Nguyen TM, Andrieu JP, Heitz A, et al. Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 2000;39:5722-30. [CrossRef]
50. Gustafson KR, McKee TC, Bokesch HR. Anti-HIV cyclotides. Curr Protein Pept Sci 2004;5:331-40. [CrossRef]
51. Tam JP, Wang S, Wong KH, Tan WL. Antimicrobial peptides from plants. Pharm 2015;8:711-57 [CrossRef]
52. Jacob B, Vogelaar A, Cadenas E, Camarero JA. Using the cyclotide scaffold for targeting biomolecular interactions in drug development. Molecules 2022;27:6430. [CrossRef]
53. Haldar UC, Saha SK, Beavis RC, Sinha NK. Trypsin inhibitors from ridged gourd (Luffa acutangula Linn.) seeds:Purification, properties, and amino acid sequences. J Protein Chem 1996;15:177-84. [CrossRef]
54. Heitz A, Hernandez JF, Gagnon J, Hong TT, Pham TT, Nguyen TM, et al. Solution structure of the squash trypsin inhibitor MCoTI-II. A new family for cyclic knottins. Biochemistry 2001;40:7973-83. [CrossRef]
55. Rodger A. Far UV protein circular dichroism. In:Encyclopedia of Biophysics. Berlin:Springer, Springer Nature;2018. 1-6. [CrossRef]
56. Kim YK, Xu H, Park NI, Boo HO, Lee SY, Park SU. Amino acid and GABA content in different cultivars of Momordica charantia L. J Med Plants Res 2009;3:894-7.
57. Hara S, Makino J, Ikenaka T. Amino acid sequences and disulfide bridges of serine proteinase inhibitors from bitter gourd (Momordica charantia LINN.) seeds. J Biochem 1989;105:88-91. [CrossRef]
58. Adewale IO, Adebiyi VG, Famutimi OG, Dada OV. Kinetics of trypsin inhibition by methanolic and solvent- partitioned fractions of two medicinal plants-Momordica charantia and Xylopia aethiopica. South Afr J Bot 2023;152:174-81. [CrossRef]
59. Dokka MK, Davuluri SP. Antimicrobial activity of a trypsin inhibitor from the seeds of Abelmoschus moschatus L. Int J Curr Microbiol Appl Sci 2014;3:184-99.
60. Ghosh I, Bhowmick BK, Jha S. Cytogenetics of two Indian varieties of Momordica charantia L.(bittergourd). Sci Hortic 2018;240:333-43. [CrossRef]