1. INTRODUCTION
More than twenty different human amyloidosis diseases and other neurodegenerative disorders have been linked to misfolding of amyloid proteins and further aggregation and fibrillation. Parkinson’s disease (PD) (a neurodegenerative disease), has been associated with pathological amyloid- α-synuclein capable of aggregation and amyloidosis. Various different types of amyloid proteins have been reported which vary in amino acid sequence but similar in their structure enriched with b-sheets giving them unique features such as robustness, stability, and insolubility in environment. These important characteristics of amyloid proteins have resulted in their selected evolution and utilization by insects, fungi, and bacteria. Thus the amyloid fiber aggregation is not just restricted to mammalian pathological and pathogenic processes but is a widely distributed attribute among the proteins and polypeptides exhibiting their existence in nature. These amyloids have been reported with biological functions such as fimbria formation in bacteria, epigenetic traits transmission in fungi and deposition and hormonal release in humans. The term “functional amyloid” has been coined to differentiate these biologically useful proteins from their toxic siblings [1,2]. Although amyloid proteins were first discovered for their link to human diseases, they now represent among a list of proteins with unique functional states which are disease-unrelated in the tree of life [3].
Recently, the microbial functional amyloids have been reported for enhancement and initiation of amyloido-genesis due to their cross-interaction. However, amyloid proteins have been reported from diverse microorganisms, their structural attributes have been studied very less. In silico studies on the proteins reduce the need for expensive laboratory work and enhance the rate of research and discovery [4,5]. Interests in the works on the gut-brain axis during the initial part of the new millennium have been reduced due to the lack of long-term longitudinal research [3]. In the present review, we have analyzed different types of bacterial “functional amyloids” with respect to α-synuclein of PD and their interaction with each other and in comparison to the known medication molecules. Understanding these interactions of “functional amyloids” of gut micro-biota with pathological (wild type and virulent) amyloid of brain influencing the mental health can help in future treatment and actions.
2. α-SYNUCLEIN – A HUMAN PATHOLOGICAL AMYLOID
In more than 50 human diseases, formation of various forms of aggregated intermediates and amyloid fibrils were observed due to misfolding and accumulation of amyloido-genic proteins [6]. In diseases such as PD (α-synuclein), Alzheimer’s disease (amyloid-β), and transmissible spongiform encephalopathy’s (prions or Platelet-rich plasma [PrP]), the cause is typically related to the structural conversions due to misfolding of a single disease associated protein [7].
Pathological aggregation of α-syn, a neuronal protein has been characterized in many progressive and pathologically similar neurodegenerative disorders such as PD, dementia with Lewy bodies (DLB), and multiple system atrophy (MSA). Both in PD and DLB, α-syn aggregation is seen as Lewy neuritis (LNs) and LBs, while in MSA it is detected as Glial cytoplasmic inclusions (GCI) within oligo-dendrocytes. Presence of these aggregates of α-syn (LBs and GCIs) with p62 and ubiquitin (of components of machinery for protein degradation) triggers the pathological cascade leading to neuro-inflammation and severe neuro-degeneration characteristic of the said diseases [8]. The wild type and misfolded α-syn are shown in Figure 1a and b.
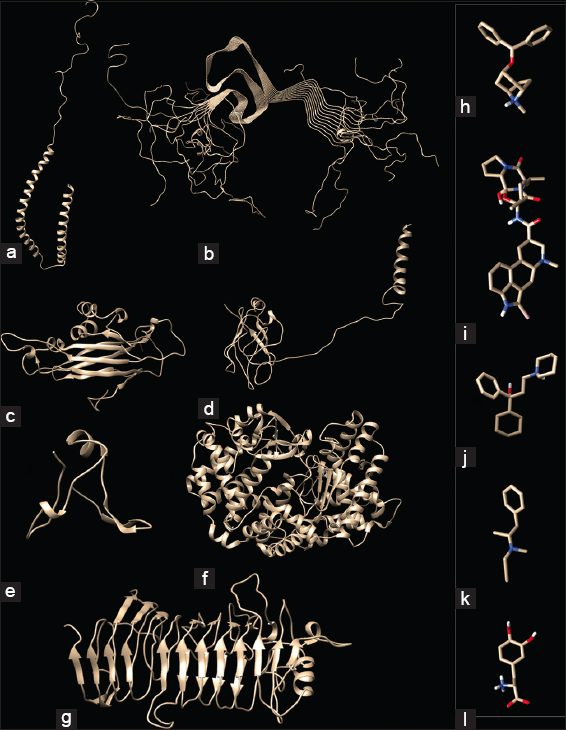 | Figure 1: Pathological and functional amyloide interactions. Structure of α-synuclein (a) Wild type (1XQ8) and (b) Misfolded (2N0A); Structure of Bacterial Functional amyloids analyzed here, (c) Bacillus subtilis (50F1) TasA amyloid, (d) Bacillus atropheus (BATR1942) TasA amyloid (e) Dinoroseobacter shibae DFL 12 (Dshi_4130), (f) Bacillus thuringiensis kurstaki YBT-1520 TasA and (g) Streptococcus pyogenes MGAS15252; Common drugs of Parkinson’s disease analyzed in the present work - (h) Benztropine, (i) Bromocriptine, (j) Trihexyphenidyl, (k) Selegiline, and (l) Levodopa. [Click here to view] |
PD, as a neurodegenerative disease shows motor and non-motor symptoms associated with decreased dopamine release and loss of dopaminergic neurons with characteristic presence of LBs or LNs. As major pathological characteristics of PD, misfolding and further deposition of α-syn was observed with related synnucleopathy and MSA. This spread of α-syn pathology across the brain region results in the onset and further progression of the clinical phenotypes. α-syn misfolding into pathologic-folds showing self-templating conformations exhibiting ability to spread between cells like “Prion” proteins and further oligomerization to form higher structures has been proposed for the pathological cascade in these neurodegenerative diseases [8-10].
3. BACTERIAL FUNCTIONAL AMYLOIDS
Amyloid fibers are ordered protein aggregates generally associated with many neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and various Prion diseases. As their production is reported from different microorganisms like, Proteobacteria, Actinobacteria, Firmicutes, and Bacteroides and also in fungi with relatively no relation to each other at the amino acid level, as seen in the present work there seems to be a preferential selection of amyloid curve during the evolution for various functions [Figure 1c-g]. The constructive use of stable amyloid fold in cellular, physiological, and functional purposes by bacteria is an evolutionary master-piece exhibiting pathological trait when similar proteins are not controlled by the cell leading to different neurodegenerative diseases. The unusual robustness of this extracellular “functional” bacterial amyloids (FuBA) (capable of withstanding chemical denaturants and SDS) has been employed by bacteria (under tight spatio-temporal-control) for biofilm-strengthening, cell-cell communications, cell-wall construction, and for bacterial antagonism. Although the misfolded human pathological amyloid proteins are intracellular-leading to cell lysis on fibrillation, most of the bacterial functional amyloids are extracellular and rarely reported for intracellular functions [3,11].
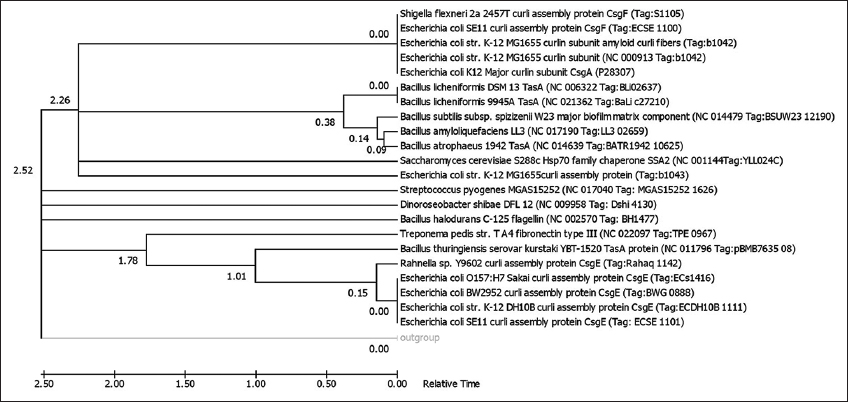
The synthesis, transport, and nucleation of bacterial FuBA involve a complex mixture of proteins makes their bioinformatic prediction and analysis with respect to pathological amyloids difficult. The increasing data piling up with genomic and protein mining can help in further research in this regard [11]. Although there have been many studies on the amyloid proto-fibril stability and oligomerization, the fibril-prone conformation and self-assembly kinetics are lacking in research data. Thus, it becomes imperative to study these transient structural oligomers in silico which can be further confirmed by lab results [12]. Many genomic and proteomic databases host a plethora of sequences for amyloids of microbial and animal origin. These functional amyloids were checked here for phylogenetic affiliation (using MEGA version 7.0 in Figure 2) and their evolutionary relationships using p-distance method, searching tree using the close-neighbor-interchange algorithm with appropriate bootstrap (5000 in Figure 2), and out-group (membrane bound Nitrosospira multiformis AmoA (AY177933) protein in Figure 2). The analysis involved 23 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 86 positions in the final dataset. Evolutionary analyses were conducted in MEGA7.0 [13-18].
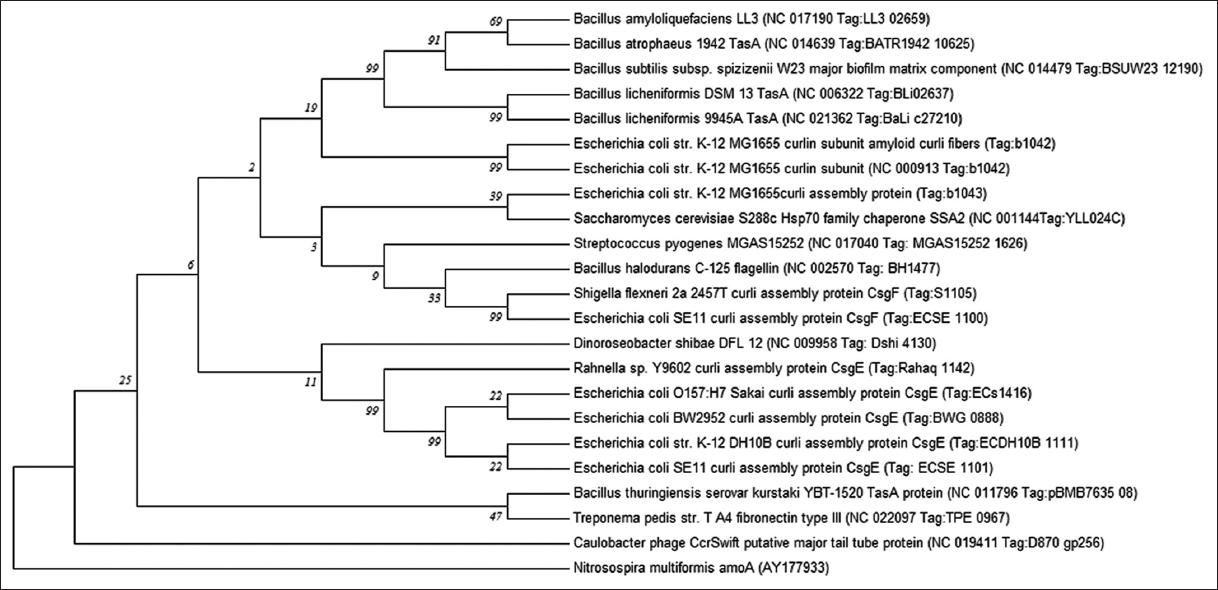 | Figure 2: Evolutionary relationships of taxa. The evolutionary history was inferred using the Minimum Evolution method [19]. The Neighbor-joining algorithm [15] was used to generate the initial tree. The optimal tree with the sum of branch length = 6.32315134 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (5000 replicates) are shown next to the branches [16]. [Click here to view] |
It can be seen through the phylogenetic tree [Figure 2], the protein homologues were non-identical even within the same genus. This fact is evident from the clustering of different proteins from Tas and Curli families belonging to Bacillus sp. and Escherichia coli sp. Respectively as the similar protein sequences did not cluster together (which can be attributed to the possible non-homologous evolution). Dino-flagellate associated Dinoroseobacter sp. and insect associated Rahnela sp. clustered with different types of curli proteins produced by strains of E. coli. Other organisms showed a relative similarity to different synthesis intermediates or subunits of E. coli curli proteins [Supplementary Table 1]. These organisms include Saccharomyces sp., Streptococcus sp., Bacillus sp., and Shigella sp. Protein homologues of Caulobacter phage clustered separately from the rest of the proteins while Bacillus thuringiensis and Treponema sp. proteins clustered together separately. Being the only phage protein homolog in the series, it was expected that the Caulobacter phage separated from the rest of the amyloid proteins.
When time tree was analyzed in present work [Figure S1], the estimated log likelihood value was −3491.9652. The analysis involved 25 amino acid sequences. All positions containing gaps and missing data were eliminated with a total of 82 positions in the final dataset. In the time tree, E. coli based curli structural proteins available in the GenBank were distinct and clustered separately into two specific clads - CsgE in a single branch while CsgA and CsgF (also Shigella flexneri CsgF) clustered together. While curli protein (Tag b1043) clad were different from the rest of related protein branches. All the Bacillus species Tas A proteins clustered together, Bacillus subtilis kurstaki YBT 1520 Tas A protein clustered with E. coli CsgE protein and Treponema pedis structural protein branch and Bacillus halodurans C-125 flagellin protein clustered as a distinct branch. The amyloid proteins from Saccharomyces cervisiae S288c, Streptococcus pyogenes MGAS15252, and Dinoroseobacter shibae DFL12 clustered as different branches from the root and outgroup. The curli proteins and Tas proteins showed distinct relative time line of evolution. While E. coli CsgE protein (surface protein) differentiated from root at 1.78, E. coli CsgA and Bacillus Species Tas A proteins (being amyloid proteins) showed a branching at 2.26 from the root. Thus, there lies the possibility that functional amyloid proteins evolved later in the bacterial system than other surface structural proteins involved in their production.
The phylogenetic analysis by the minimum evolution tree here shows differences among the protein structures of different microbial amyloids. Nielsen et al. [20] describe this fact of amyloid variation with respect to their distribution across the living kingdom as “each species, its own amyloid.”
When Teoh et al. [21] analyzed the protein sequences of apo-lipoprotein superfamily, a high level of conservation was revealed. This was assumed to be due to duplications of ancestral amino acid tandem repeats forming a number of structural features in the common background of the proteins. Thus indicating that during the evolution for various functions, the amyloid fold has been selected multiple times. As seen in a newly described class of FuBA, the amyloid formation can be an integral part of normal cellular physiology. Giraldo [3] studied the subtle differences among these functional amyloids with respect to pathogenic and transmissible amyloids especially RepA-WH1, a bacterial amyloidogenic functional protein controlling the plasmid replication [2].
4. GUT MICROFLORA AND AMYLOID BIOSYNTHESIS
During the evolutionary process, humans have developed a sturdy symbiotic relationship with their gut micro-biome consisting of a complex community of archaea, bacteria, protists and viruses (including bacteriophages). Each individual harbors unique set of gut bacteria that influence interpersonal heterogeneity. This micro-biota enhances the selective neurological disorders due to the metabolism of polyphenols that interfere with the key pathologic mechanisms. The accumulation of these flavonoid metabolites have the ability to interact with α-syn and play a major role in their misfolding, inflammations, α-synucleiopathies, and PD progression [21].
The conformational variation in Pseudomonad amyloid FapC can be used as a potential therapeutic target to understand the bacterial colonization and infection during Alzheimer’s disease [22]. Many bacteria among the mouth micro-flora have been reported for release of FuBA and extensive cases have been reported from Streptococcus mutans and other bacteria involved in periodontal infections [23-25]. Goya et al. [27] reported effect of B. subtilis strain PXN21 vegetative cells and spores as psychobiotics in inhibiting α-syn and synucleiopathy and recommend their use as dietary supplements.
5. CROSS REACTIVITY OF BACTERIAL AMYLOIDS WITH A-SYNUCLEIN
Amyloido-genesis has been known to be influenced by genetic and environmental factors. Recent evidences have suggested the acceleration of amyloid aggregation due to non-identical amyloid proteins as seen in prion proteins due to possible cross-interactions between heterologous amyloidogenic proteins leading to amyloidogenesis and related disease pathology [7,27,28].
In recent years, lot of attention has been paid to the gut-brain axis with increasing data as symptomatic, physiological, and pathological findings accumulated [22]. The gut-brain bidirectional communication is controlled by the vagus nerve (VN) with the help of enteric nervous system (ENS). The neuro-pathological characteristic of PD is the widespread α-synucleiopathy in central and peripheral nervous systems including in ENS. The initiation of α-syn aggregation have also been linked to bacterial exudates of gut micro-biota in the ENS, which can then be transmitted to the CNS through VN in a prion-like manner [29]. Resident microbial community of the individual as micro-biota especially those capable of producing bacterial amyloids and other amphipathic metabolites have been known to influence the α-syn aggregation and influence the development of PD [30]. Microbial interaction with the VN and a similar cascade of event as given for neuro-pathological diseases has been reported for the action of psychobiotics in enhancing the mental health [31].
As seen in Figure 3, the interaction between bacterial amyloids with wild type α-synuclein resulted in specific regions being marked for repeated interactions [Figure 3a-e], which was correlating with the interaction potential of misfolded α-synuclein [Figure 3f]. Thus, the bacterial amyloids have the capability to interact with wild type α-synuclein and increase or decrease the progression of the PD in patients.
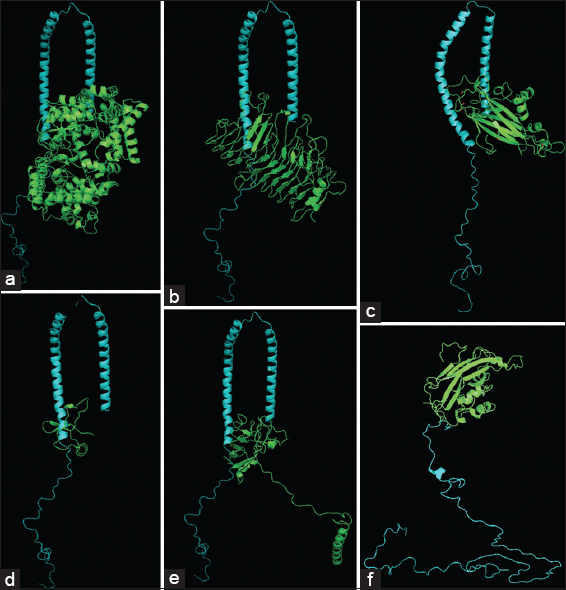 | Figure 3: Interaction of α-synuclein with bacterial functional amyloids. Docking studies for α-synuclein (1XQ8) with (a) Streptococcus pyogenes MGAS15252, (b) Dinoroseobacter shibae DFL 12 (Dshi_4130), (d) Bacillus thuringiensis kurstaki YBT-1520 TasA, (e) Bacillus atropheus (BATR1942) TasA and B. subtilis (50F1) TasA amyloid (c and f). [Click here to view] |
Various bacterial functional amyloids are allowed in present work to interact with α-synuclein by docking studies using Autodock Vina and using GROMACS software and molecular dynamics (MD) simulations up to 100 ns for analyzing the interaction stability. As shown in supplementary Figure 2, the bacterial functional amyloids were able to form stable connections with wild type α-synuclein and the major interacting amino-acids seen to be interacting with PHE4, LYS97, MET5, and ALA90 consistently while for B. subtilis (50F1) TasA amyloid, interaction at THR137 consistently even with misfolded α-synuclein [4]. The mid protein interaction of BaFu exhibited their effects on protein folding of α-synuclein which converts to misfolded pattern in 60 ns when checked in MD simulation studies [Supplementary Figure 2].
Although α-synuclein aggregates are considered prion-like for their communicable form of disease progression, the preclinical models of disease progression are facing the limitations of proof of causality especially when there exists confounding effects of like long incubation periods, host factors such as inflammation and immune response [32]. Werner et al. [29] reported the possible role of gut-microbiome (CsgA from E. coli) in initiation, modulation or enhancement of human neurodegenerative disease (α-synuclein causing PD). They also reported the possibility of in vitro cross-reactivity and the phenomenon as widespread among the other proteins. Sampson et al. [28] reported the enhancement of pathology in brain of α-synuclein over-expressing mice when infection and colonization of curli (bacterial functional amyloid) producing E. coli. The aggregation of α-synuclein was found to be accelerated when these mice were treated with purified Curli subunits. The amyloidogenic CsgA protein from E. coli biofilms has been reported to trigger PD in mice [28]. Nerius et al. [34] investigated the pathogenesis of PD and role of common GI infections and the most frequent among those are the causative agents of colitis and gastroenteritis followed by viral intestinal infections. The studies by Yamasaki et al. [10] suggest the possible interaction between gut microbiome and certain dietary factors which can be used as an effective modulators for pathologic propagation of α-syn. Olsen [25] reported the link between the biomolecules (amphi-philes and proteins) released by Peridontotitis causative Porphyromonas gingivalis in enhancing the systemic inflammation and neuro-inflammation seen in AD and PD.
The evidences suggest that the α-syn inclusions undergo template directed aggregation propagation (template-directed) among the brain and peripheral organs in a prion-like manner. Reports of integral loop of transmission of α-syn as a prionoid from periphery to the brain via specific pathways [9]. It is important to analyze and investigate other aspects of cross-reactivity among the bacterial functional amyloids and human pathological amyloids (α-synuclein) to understand the underlying mechanisms and to develop new treatments depending on the altered gut microbiome [28].
25 major bacterial functional amyloid sequences were collected (from GenBank) and were used to build homology models (using Swissmodel, Phyre2 and RAPTOR-X servers and analyzed for protein parameters on Protparam site) [4]. The models were submitted to PMDB [Supplementary Table 1] and five among the models were selected for further analysis. The models were chosen according to their presence in the time tree clustering. Other than Curli and Tas A proteins from E. coli (E. coli K12 CsgA protein [P28307]) and Bacillus species (B. thuringiensis serovar kurstaki YBT-1520 (PBMB7635_08) and Bacillus atropheus 1942 (BATR1942_10625)), respectively, amyloid models were chosen from S. pyogenes MGAS15252 (MGAS15252_1626) and D. shibae DFL12 (Dshi_4130). The five selected amyloids were further analyzed by the MD simulation studies. Computational modeling is a tool that can complement and predict the time consuming and expensive in vitro experiments on the stability of amyloid oligomers and their assembly. MD simulation studies have potential to identify the stability determining factors of amyloid proteins oligomers [34]. In GROMACS, the protein dimensions can be treated by considering a series of physiological parameters. This can be used for MD simulation of the effects on the protein molecule [4,5].
6. CROSS-REACTIVITY OF DRUGS OF MISFOLDED A-SYNUCLEIN AGAINST WILD TYPE PROTEIN
Drugs are made to interfere and target the sites of misfolded proteins and also hamper the nucleation of proteins in the cells reducing the burden of pathological trait of α-syn aggregation leading to synucleiopathies [8]. Some of the drugs which have been analyzed in the present review are given in Figure 1h-l.
Antiparkinsonian agents such as bromocriptine, levodopa, benztropine, trihexyphenidyl, and selegiline were docked against native (non-misfolded) α-synuclein and analyzed for interaction stability using MD simulation (up to 100 ns) (in present study) [Figure 4a-e]. The interactions were found to be mirroring the interactions by bacterial functional amyloids except Trihexyphenidyl which had a weaker interaction at the THR137. Pretorius et al. [36] reported the binding of lipopolysaccharide binding protein in controlling the nucleation and amyloid fiber elongation in PD.
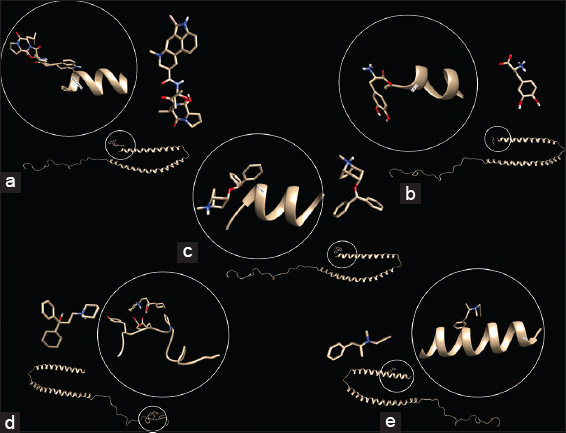 | Figure 4: In silico interaction studies between α-synuclein and anti-Parkinsonian drugs. Interaction of wild type α-synuclein (1XQ8) with (a) Bromocriptine, (b) Levodopa, (c) Benztropine, (d) Trihexyphenidyl, and (e) Selegiline using AutoDock Vina. [Click here to view] |
Many anti-Parkinsonian agents (levodopa, selegiline, and bromocriptine) have been reported to exhibit in vitro inhibition of photo-induced cross-linking of unmodified amyloid proteins involved in neuro-degeneration (Alzheimer’s and PD), while some (Trihexyphenidyl) could not interact with these proteins and could be involved with other pathways for Parkinson’s treatment [36]. Shirasaka et al. [7] reported the amyloidogenesis of α-synuclein without interaction with monomeric or fibrillar prion protein (PrP) nucleation domains, thus indicating the possible interactions (by binding) at partially unfolded state of transiently accumulated intermediate. Thus the interactions of microbial functional amyloids have the additional ability to act as anti-Parkinsonian drugs and thus can enhance mental health. This phenomenon has been utilized in application of such microbes as psychobiotics [31].
7. PSYCHO-DIETARY EFFECTS ON THE NEURO-DEGENERATIVE DISEASES
The causal effects of gut micro-biota on emotional experience particularly stress, depression and anxiety in patients enhanced the focus of the researchers on their contributions there has been enhanced studies dealing with neuroscience and biopsychology of interaction in the axis of brain-gut-microbiome [37]. Recent studies have revealed the use of this gut microbiome-brain axis in modifying cognitive and behavioral properties of patients using live beneficial microorganisms termed “Psychobiotics.” Here, the introduction of these bacteria can either engage directly one or more gut-brain circuits or to alter the established gut micro-biota of the recipient modulating the signaling to the brain [38].
Elucidation of the effect of introduced gut-micro-biota on the germ-free laboratory animals (Mice) showed the increased effects on the function of hippocampus and amygdala resulting in effects on short-term recognition, working memory and display of altered social behavior [37]. Studies have found associations of microbiome function and composition influencing the neuro-behavioral and affective disorders including parameters of neuroimaging. The microbial re-colonization with respect to the age of the patients shows time-sensitive efficacy for microbial reconstitution with possibility of critical window for neurodevelopment within which abnormalities arising due to the absent micro-biota can be reversed by time dependent microbial re-colonization [38].
The uncertainties about the clinical applications of psychobiotics persists as the mechanics of action have not been identified yet and the time period as well as durability of their usage. The reports of changes in responses to psychobiotics in a wide range of brain regions have been questioned for possible inadequate questionnaires used for short-term studies. Effects of psycho-biotic-intervention may also depend on the pre-existing microbiome and neuro-anatomy, diet as well as genetics of the recipients. The studies on their clinical interventions in humans and corresponding effects on emotion, cognition and behavior have been met with scepticism due to the worldwide use of psycho-biotic microorganisms as probiotics without significant effects on psychological traits [37,38]. Various microorganisms breaching the gut-brain barrier and influencing the health of individual in positive and negative ways are listed in Table 1.
Table 1: Effects of reported microorganisms on the brain health during Parkinson’s disease.
| S. No. | Organism | Product | Interaction | References |
|---|---|---|---|---|
| Negative | ||||
| 1 | Gut microbiome | Flavonoid metabolites | Interact with α-syn and influence their misfolding, inflammations, α-synucleiopathies and PD progression | Ho et al. [22] |
| 2 | Pseudomonas sp. | FapC | Bacterial colonization and infection during Alzheimer’s disease | Javed et al.[23] |
| 3 | Streptococcus mutans and other bacteria involved in periodontal infections | Amyloids | Interact with α-syn and PD progression | Paranjapaye and Daggett; Ranjan et al; Olsen, [24-26] |
| 4 | E. coli | CsgA | Trigger and enhance the progression of PD | Werner et al. [29] |
| 5 | Faecalibacterium species | Butyrate, lipopolysaccharide and other secondary metabolite biosynthesis | colonic inflammation and PD progression with enhanced a-Syn aggregation | Keshavarzian et al. [40] |
| 6 | D. desulfuricans MB (DSM 6949) and D. vulgaris DSM 644 | Dissimilatory sulphate reduction and production of hydrogen sulfide (H2S) & magnetite. | Uncoated magnetite nanoparticles can accelerate a-Syn aggregation | Murros [41] |
| 7 | Staphylococcus aureus | Functional Amyloids-Modulins | Catalyze Alpha-Synuclein Aggregation | Haikal et al. [42] |
| 8 | Escherichia coli | Amyloid Curli | Promotes α-synuclein aggregation and motor impairment in mice | Sampson et al. [28] |
| Positive | ||||
| 9 | Faecal microbiota of healthy donors | Short chain fatty acids (SCFAs) and lactate | Control of progression of PD | Ghyselinck et al. [43] |
| 10 | Bacillus subtilis strain PXN21 | Vegetative cells and spores | Psychobiotics in inhibiting α-syn and synucleiopathy | Goya et al. [27] |
| 11 | Lactic acid bacteria | Metabolites crossing gut-brain barrier | Psychobiotics | Toro-Barbosa et al.; Castelli et al. [32,44] |
| 12 | Lactobacillus plantarum DP189 | Improved behavioral ability and increased levels of 5-hydroxytryptamine and dopamine of PD | Neuro-protective and Control of progression of PD | Wang et al. [45] |
| 13 | Salmonella typhimurium VNP20009 (ATCC 202165) construct MG1363-pMG36e-GLP-1 | Glucagon-like peptide-1 (GLP-1) | Neuro-protective effects in Parkinson disease mouse model | Chen et al.; Fang et al. [49,50] |
| 14 | Gut microbiota with elevated Bacteroidetes and fermicutes phyla | Osteocalcin protein | Modulate brain function and exert protective effect on PD | Hou et al. [48] |
Greater insights into the mechanisms including molecular factors that drive interactions between the microbiome of the gut, human brain function and psychology can be established by studying the brain-gut microbiome axis using “bottom up” approach. For progress in analyzing the brain-gut micro-biota axis needs multidisciplinary work elaborating the host-microbe interactions which can further develop our knowledge about the effects of this axis on the emotions and information processing as well as its interaction in context to cultural behavior [37].
8. CONCLUSION AND FUTURE PERSPECTIVES
The specific communication channels between the gut micro-biota and brain have already been elucidated to include - vagal nerve signaling, enteric nerve activation, entero-endocrine, and entero-chromaffin cells stimulation and the release of neuro-active products into circulation leading to modulation of neuro-immune response. As seen in the present review, the “functional amyloids” of microbial origin have the sites for interaction with wild type α-synuclein which are similar to the interaction of anti-Parkinsonian drugs. Thus these sites of interaction can be utilized for enhancement or control of amyloidosis and fibrillation in patients with PD earning the suitable name of – “Psychobiotics.” Further in vitro and in vivo studies are necessary for tapping the potential of psychobiotics and their “functional amyloids” for controlling PD. Elucidating the effects of psychological developments as well as cultural and social factors on the brain-gut microbiome axis can help for contextualizing the role of this axis and corresponding psychological interventions to improve the human health.
9. ACKNOWLEDGMENTS
The authors would like to acknowledge the help of Heads and Principals of their respective colleges.
10. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
11. FUNDING
The authors declare that no funding was received for the present work.
12. CONFLICTS OF INTEREST
The authors declare that there is no financial or any other conflicts of interest in this work.
13. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
14. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
15. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Markande AR, Nerurkar AS. Microcosm-based interaction studies between members of two ecophysiological groups of bioemulsifier producer and a hydrocarbon degrader from the Indian intertidal zone. Environ Sci Pollut Res 2016;23:14462-71. [CrossRef]
2. Markande AR, Nerurkar AS. Analysis of nutritional factors influencing the biosynthesis of amyloid bioemulsifier BE-AM1 applicable in food industry. J Microbiol Biotechnol Food Sci 2021;11:e3170. [CrossRef]
3. Giraldo R. SynBio and the boundaries between functional and pathogenic RepA-WH1 bacterial amyloids. mSystems 2020;5:e00553-20. [CrossRef]
4. Mehta K, Markande A.
5. Mehta K, Markande A.
6. Jia L, Zhao W, Wei W, Guo X, Wang W, Wang Y,
7. Shirasaka M, Kuwata K, Honda R. a-Synuclein chaperone suppresses nucleation and amyloidogenesis of prion protein. Biochem Biophys Res Commun 2020;521:259-64. [CrossRef]
8. Fouka M, Mavroeidi P, Tsaka G, Xilouri M. In search of effective treatments targeting a-synuclein toxicity in synucleinopathies:Pros and cons. Front Cell Dev Biol 2020;8:559791. [CrossRef]
9. Zheng H, Shi C, Luo H, Fan L, Yang Z, Hu X,
10. Yamasaki TR, Ono K, Ho L, Pasinetti GM. Gut microbiome-modified polyphenolic compounds inhibit a-synuclein seeding and spreading in a-synucleinopathies. Front Neurosci 2020;14:398. [CrossRef]
11. Christensen LF, Schafer N, Wolf-Perez A, Madsen DJ, Otzen DE. Bacterial amyloids:Biogenesis and biomaterials. Adv Exp Med Biol 2019;1174:113-59. [CrossRef]
12. Nguyen PH, Li MS, Derreumaux P. Effects of all-atom force fields on amyloid oligomerization:Replica exchange molecular dynamics simulations of the Ab 16-22 dimer and trimer. Phys Chem Chem Phys 2011;13:9778-88. [CrossRef]
13. Kumar S, Stecher G, Tamura K. MEGA7:Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-4. [CrossRef]
14. Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York:Oxford University Press;2000.
15. Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-25.
16. Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution 1985;39:783-91. [CrossRef]
17. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5:Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-9. [CrossRef]
18. Tamura K, Battistuzzi FU, Billing-Ross P, Murillo O, Filipski A, Kumar S. Estimating divergence times in large molecular phylogenies. Proc Natl Acad Sci U S A 2012;109:19333-8. [CrossRef]
19. Rzhetsky A, Nei M. A simple method for estimating and testing minimum-evolution trees. Mol Biol Evol 1992;9:945-67.
20. Nielsen PH, Dueholm MS, Thomsen TR. Functional bacterial amyloids in biofilms. In:Flemming HC, Szwezyk U, Wingender J, editors. Annual Biofilm Highlights. Berlin Heidelberg:Springer-Verlag;2011. 41-62. [CrossRef]
21. Teoh CL, Pham CL, Todorova N, Hung A, Lincoln CN,
22. Ho L, Zhao D, Ono K, Ruan K, Mogno I, Tsuji M,
23. Javed I, Zhang Z, Adamcik J, Andrikopoulos N, Li Y, Otzen DE,
24. Paranjapye N, Daggett V.
25. Ranjan R, Dhar G, Sahu S, Nayak N, Mishra M. Periodontal disease and neurodegeneration:The possible pathway and contribution from periodontal infections. J Clin Diagn Res 2018;12:DE01-5. [CrossRef]
26. Olsen I. Possible link between
27. Goya ME, Xue F, Sampedro-Torres-Quevedo C, Arnaouteli S, Riquelme-Dominguez L, Romanowski A,
28. Sampson TR, Challis C, Jain N, Moiseyenko A, Ladinsky MS, Shastri GG,
29. Werner T, Horvath I, Wittung-Stafshede P. Crosstalk between alpha-synuclein and other human and non-human amyloidogenic proteins:Consequences for amyloid formation in Parkinson's disease. J Parkinsons Dis 2020;10:819-30. [CrossRef]
30. Dos Santos CI, França YR, Campos CD, Bomfim MR, Melo BO, Holanda RA,
31. Miraglia F, Colla E. Microbiome, Parkinson's disease and molecular mimicry. Cells 2019;8:222. [CrossRef]
32. Del Toro-Barbosa M, Hurtado-Romero A, Garcia-Amezquita LE, García-Cayuela T. Psychobiotics:Mechanisms of action, evaluation methods and effectiveness in applications with food products. Nutrients 2020;12:3896. [CrossRef]
33. Leak RK, Frosch MP, Beach TG, Halliday GM. Alpha-synuclein:Prion or prion-like?Acta Neuropathol 2019;138:509-14. [CrossRef]
34. Nerius M, Doblhammer G, Tamgüney G. GI infections are associated with an increased risk of Parkinson's disease. Gut 2020;69:1154-6. [CrossRef]
35. Berhanu WM, Hansmann UH. Structure and dynamics of amyloid-b segmental polymorphisms. PLoS One 2012;7:e41479. [CrossRef]
36. Pretorius E, Page MJ, Mbotwe S, Kell DB. Lipopolysaccharide-binding protein (LBP) can reverse the amyloid state of fibrin seen or induced in Parkinson's disease. PLoS One 2018;13:e0192121. [CrossRef]
37. Ono K, Takasaki JI, Takahashi R, Ikeda T, Yamada M. Effects of antiparkinsonian agents on b-amyloid and a-synuclein oligomer formation
38. Allen AP, Dinan TG, Clarke G, Cryan JF. A psychology of the human brain-gut-microbiome axis. Soc Personal Psychol Compass 2017;11:e12309. [CrossRef]
39. Jacobs JP, Mayer EA. Psychobiotics:Shaping the mind with gut bacteria. Am J Gastroenterol 2019;114:1034-5. [CrossRef]
40. Keshavarzian A, Green SJ, Engen PA, Voigt RM, Naqib A, Forsyth CB,
41. Murros KE, Huynh VA, Takala TM, Saris PE.
42. Haikal C, Ortigosa-Pascual L, Najarzadeh Z, Bernfur K, Svanbergsson A, Otzen DE,
43. Ghyselinck J, Verstrepen L, Moens F, Van Den Abbeele P, Bruggeman A, Said J,
44. Castelli V, d'Angelo M, Lombardi F, Alfonsetti M, Antonosante A, Catanesi M,
45. Wang L, Li S, Jiang Y, Zhao Z, Shen Y, Zhang J,
46. Li C, Qin R, Liu R, Miao S, Yang P. Functional amyloid materials at surfaces interfaces. Biomater Sci 2018;6:462-72. [CrossRef]
47. Xin XF, Kvitko B, He SY.
48. Hou YF, Shan C, Zhuang SY, Zhuang QQ, Ghosh A, Zhu KC,
49. Chen G, Tang B, Yang BY, Chen JX, Zhou JH, Li JH,
50. Fang X, Zhou X, Miao Y, Han Y, Wei J, Chen T. Therapeutic effect of GLP-1 engineered strain on mice model of Alzheimer's disease and Parkinson's disease. Amb Express 2020;10:1-3. https://doi.org/10.1186/s13568-020-01014-6 PMid:32333225 PMCid:PMC7182653
SUPPLEMENTARY MATERIALS
Supplementary Table 1: The homology proteins generated and their PMDB ID.
| S. No. | Organism name | Tag | PMDB ID |
|---|---|---|---|
| 1 | Bacillus amyloliquefaciens LL3 | LL3_02659 | PM0080881 |
| 2 | Bacillus atrophaeus 1942 TasA | BATR1942_10625 | PM0080907 |
| 3 | Bacillus halodurans C-125_flagellin | BH1477 | PM0080908 |
| 4 | Bacillus licheniformis 9945A_TasA | BaLi_c27210 | PM0080882 |
| 5 | Bacillus licheniformis DSM 13_TasA | Bli02637 | PM0080883 |
| 6 | Bacillus subtilis spizizenii W23 major biofilm matrix component | BSUW23_12190 | PM0080884 |
| 7 | Bacillus thuringiensis kurstaki YBT-1520_TasA | pBMB7635_08 | PM0080885 |
| 8 | Caulobacter phage CcrSwift | D870_gp256 | PM0080886 |
| 9 | Dinoroseobacter shibae DFL_12 | Dshi_4130 | PM0080887 |
| 10 | Escherichia coli BW2952 curli assembly protein CsgE | BWG_0888 | PM0080888 |
| 11 | Escherichia coli K12 DH10B curli assembly protein CsgE | ECDH10B_1111 | PM0080889 |
| 12 | Escherichia coli (strain K12)_CsgA_ | P28307 | PM0080890 |
| 13 | Escherichia coli K-12 MG1655 curli assembly protein | S1105 | PM0080891 |
| 14 | Escherichia coli K-12 MG1655 curlin | NC_000913 | PM0080892 |
| 15 | Escherichia coli K-12 MG1655 curlin subunit | b1042 | PM0080893 |
| 16 | Escherichia coli O157H7 Sakai curli assembly protein CsgE | NC_002695.1 | PM0080894 |
| 17 | Escherichia coli K-12 MG1655 curlin | NC_000913.3 | PM0080897 |
| 18 | Escherichia coli K-12 MG1655 curlin subunit | b1042 | PM0080898 |
| 19 | Escherichia coli O157H7 Sakai curli assembly protein CsgE | NC_002695.1 | PM0080899 |
| 20 | Escherichia coli SE11 curli assembly protein CsgF | ECSE_1100 | PM0080900 |
| 21 | Escherichia coli SE11 curli assembly protein CsgE | ECSE_1101 | PM0080901 |
| 22 | Rahnella sp. Y9602 curli assembly protein CsgE | Rahaq_1142 | PM0080902 |
| 23 | Saccharomyces cerevisiae S288c Hsp70 | YLL024C | PM0080903 |
| 24 | Streptococcus pyogenes MGAS15252 | MGAS15252_1626 | PM0080904 |
| 25 | Treponema pedis TA4 fibronectin typeIII | TPE_0967 | PM0080905 |
 | Supplementary Figure 1: Time tree of bacterial functional amyloids. [Click here to view] |
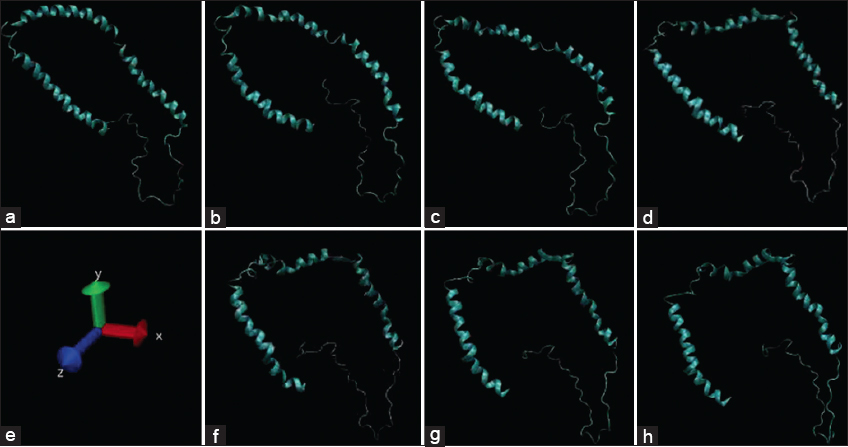 | Supplementary Figure 2: Misfolding of α-synuclein to amyloid folds in first 40 ns under molecular dynamics simulations. [Click here to view] |