Heavy metals (HMs) are typically classified as metallic elements and metalloids with high molecular weight and density above 5 g/cm3 [1]. In contrast to organic pollutants, HM pollutants cannot be naturally eliminated and begin to bioaccumulate in sediments, subsequently entering the food chain. In addition, the buildup of potentially harmful HMs can adversely affect the physical metabolism, activity, biomass, diversity, and composition of plants and microbial communities [2]. Chromium (Cr) is toxic to plants, microorganisms, and animals at higher concentrations [3,4]. Cr (III) and Cr (VI) compounds are classified as Group 3 and Group 1, respectively, by the International Agency for Research on Cancer [5]. Sources of chromium include both natural processes, such as the weathering of parent rocks and volcanic eruptions, and anthropogenic activities, including fossil fuel combustion, industrial operations, chromium plating, leather tanning, metal fabrication, wood preservatives, pulp and paper mills, paints, printing inks, and anti-corrosive materials [6]. Among the various forms of chromium (Cr+6, Cr+5, Cr+4, Cr+3, Cr+2, Cr+1, Cr0, Cr−1, and Cr−2), Cr (VI) and Cr (III) are the most stable. The excessive accumulation of Cr (VI) in the soil poses significant challenges to plant growth and crop productivity [7].
Addressing HM contamination is essential for maintaining agricultural soil health, as it directly affects crop growth and productivity. Recently, various physical and chemical methods have been employed to remediate contaminated soils. While the use of chelators is integral to these methods, their excessive application can pollute soil and groundwater. In addition, these traditional methods are non-eco-friendly and expensive and require skilled operators for the machinery involved [8]. The use of bacteria in phytoremediation has been widely accepted because of their significant impact on plant growth and their ability to enhance the remediation efficiency of contaminated soils [9]. Plant-microbe interactions are a vital approach to addressing HM contamination in soil. Microorganisms such as bacteria, protozoa, fungi, and algae inhabit the soil around plant roots, known as the rhizosphere. Plant growth-promoting bacteria/rhizobacteria (PGPB/R) are also found in the rhizosphere. These PGPBs naturally associate with plants in various ways, functioning as free-living, symbiotic, or endophytic organisms [10]. Among microbes, metal-resistant plant growth-promoting rhizobacteria (PGPR) are promising candidates for biofertilization and bioremediation, reducing the reliance on chemical fertilizers. Moreover, these microbes can mitigate both abiotic and biotic stresses [11]. In addition, certain microorganisms may be crucial in the biogeochemical cycling of toxic HMs and the remediation of metal contamination. Numerous studies have investigated the potential of microorganisms in mitigating HM contamination, with some yielding promising results [12]. PGPR also mitigates salt stress and induces drought tolerance in host plants by altering root system architecture, ion transport system, aiding osmoregulation, exopolysaccharides (EPS) activity, biofilm formation, activating antioxidant defense systems to scavenge reactive oxygen species (ROS), reducing lipid peroxidation, regulating phytohormones, and transcriptionally regulating host stress-responsive genes [13,14].
Numerous bacterial genera have been identified as PGPB, including Agrobacterium, Alcaligenes, Arthrobacter, Azospirillum, Azotobacter, Bacillus, Burkholderia, Clostridium, Enterobacter, Klebsiella, Phyllobacterium, Pseudomonas, Rhizobium, Serratia, and Xanthomonas [15]. Studies showed that plant growth improves under both metal-exposed and unexposed conditions, either through biocontrol activities or by enhancing various plant growth promotion (PGP) traits such as the production of 1-aminocyclopropane-1-carboxylate deaminase, hydrogen cyanide, siderophores, indole-3-acetic acid, nitrogen fixation, ammonia production, and phosphate solubilization [16]. Due to the direct and indirect effects of PGPR on host plants, they are ideal candidates for commercialization as bioinoculants; thereby contributing to the goals of sustainable agriculture [17]. However, researchers are actively seeking new strains of PGP and biocontrol endophytic bacteria to enhance plant growth and reduce reliance on agrochemicals. The utilization of PGP endophytes that enhance plant growth and nutrient uptake in nutrient-deficient conditions represents a significant step toward sustainable agriculture. Recent studies have extensively documented the role of endophytic bacterial strains in the biofortification of various crop plants [18]. Biosorption is a process where living and non-living microbial cells, along with cellular products such as polysaccharides, are utilized to remove HM ions from aqueous solutions based on adsorption principles. This bioremediation technique has garnered significant attention in recent years [19]. Numerous microorganisms have been genetically engineered to enhance their capacity for HM bioremediation. Engineered strains such as Escherichia coli BL21, E. coli TOP10, E. coli MC4100, B. subtilis PY79, E. coli BL21 (DE3), and R. palustris have demonstrated effective bioremediation of lead (Pb), cadmium (Cd), copper (Cu), nickel (Ni), arsenic (As), and mercury (Hg), respectively [20]. However, concerns regarding the accidental release of these modified organisms have limited their widespread application [12]. This study aimed to investigate endophytic bacteria isolated from the local region of West Bengal, India, and to evaluate their PGP attributes under varying concentrations of Cr (VI) stress.
2. MATERIALS AND METHODS
2.1. Collection, Isolation, and Maintenance of Endophytic Bacteria
Healthy chickpea (Cicer arietinum) plants were collected from an agricultural field located near a village area named Krishipally, block Santipur, and district Nadia region of West Bengal, India (latitude 23° 14’ 24.733” N; longitude 88°3’ 25.789” E). The plants were kept in a ziplock bag for transport to the laboratory. In the laboratory, the root parts were carefully separated from the shoot parts. The plant root parts were thoroughly washed with tap water for 5 min, followed by three subsequent washes with sterile-distilled water. The nodules were carefully excised from the root, followed by washing twice with 70% ethanol (v/v) for 3 min and then sterilized with 0.1% mercuric chloride (w/v) for 2 min. Samples were then finally washed thrice using sterile distilled water. After surface sterilization, each nodule was placed in a sterile 1.5 mL centrifuge tube, and 500 µL of sterile water was added. The nodule was then crushed using an autoclaved plastic homogenizer. The 500 µL of nodular sap was serially diluted up to 10–4 times, followed by the spreading of 100 µL diluted sap on yeast mannitol agar containing Congo red (YMACR) (HiMedia M721, India). Further, the plates were incubated at 28 ± 2°C for 24–48 h. After incubation, the plates were observed for morphologically distinct colonies. Fifteen distinct isolates were observed, then carefully sub-cultured onto YMACR to obtain pure cultures. Moreover, 25% (v/v) glycerol stocks were prepared from fresh overnight cultures for long-term storage at −80°C.
2.2. Screening of Chromium-Tolerant Bacteria
Chromium-tolerant endophytic bacteria were screened using the gradient Petri plate method [21]. A two-layer agar method was used in 125 mm square Petri dishes measuring 120 × 120 × 15 mm (Himedia, Cat. No. PW050), with a maximum volume capacity of approximately 216 mL, to create linear gradient plates with increasing chromium concentrations. The bottom layer consisted of 100 mL of yeast mannitol agar supplemented with 500 µM Cr (VI) (K2Cr2O7), which was allowed to solidify in a tilted position to form a slant to ensure even coverage across the bottom. Once solidified, the plate was placed horizontally, and 100 mL of plain yeast mannitol agar was poured as the top layer. The plates were left undisturbed for 12 h at room temperature (~25°C) to allow vertical diffusion of Cr (VI) from the bottom layer. This setup generated a linear gradient with Cr (VI) concentrations ranging from 0 to 500 µM along the gradient axis. The plate was divided into six grids, where bacterial isolates were spot-inoculated to grow up to the final 6th grid in the plate gradient assay. In this assay, isolates were inoculated onto six successive grids representing increasing concentrations of chromium (0–500 µM). Isolates that exhibited visible growth across the highest number of grids were considered more tolerant to Cr (VI) stress and thus demonstrated superior performance relative to others. This method enabled the evaluation of bacterial tolerance to Cr (VI) across a gradient of HM stress. Fifteen bacterial isolates were screened for Cr (VI) tolerance, of which two exhibited superior performance compared to the others. These two isolates were designated as strains SMAJ_44 and SMAJ_47 [Figure 1]. Moreover, subsequent detailed morphological and biochemical tests were conducted on these two strains for their phenotypic characterization according to Bergey’s Manual of Determinative Bacteriology [22].
 | Figure 1: Gradient plate assay with two agar layers containing increasing concentrations of Cr (VI) (0–500 µM) to assess bacterial tolerance using spot assay. [Click here to view] |
2.3. Molecular Identification of the Bacterial Strains
2.3.1. Genomic DNA extraction
The two bacterial strains SMAJ_44 and SMAJ_47 were grown in 2 mL yeast mannitol broth (HiMedia M716, India), pH 7.0, followed by incubation for 24 h at 28 ± 2°C, with constant shaking at 100 rpm. Then the culture was centrifuged at 10,000 × g for 5 min at 4°C (Eppendorf Centrifuge 5424R) to obtain the cell pellet, while the supernatant was discarded. Genomic DNA was extracted from the pellet, using the HiPurA® Bacterial Genomic DNA Purification Kit (Himedia MB505, India) [23].
2.3.2. Polymerase chain reaction (PCR) and 16S rDNA amplification
For molecular identification and characterization, the genomic DNA of two bacterial strains, PCR amplification was performed targeting the 16S rRNA gene for molecular identification of the bacterial isolates using the universal primers 27F 5’-AGAGTTTGATCGTGGCTCAG-3’ and 1492R 5’-CGGTTACCTTGTTACGACTT-3’ purchased from IDT (Integrated DNA Technologies) [24]. Both the forward and reverse primers were added to the 50 µL of reaction mixture containing 1× reaction buffer including 1.5 mM MgCl2, 0.2 µM of each of the forward and reverse primers, 0.2 mM dNTP, 5 µL of the template DNA, water to a final volume of 50 µL, and 1.25 U of Taq DNA polymerase. The following PCR program was used: An initial denaturation at 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 60 s, and a final extension at 72°C for 10 min. All of the aforementioned chemicals were procured from BioBharati LifeScience Pvt. Ltd. (Kolkata, India) [25]. A sample containing sterile water instead of DNA was used as a negative control. The PCR reactions were carried out in a T 100™ Thermal cycler (BioRad). The size of the amplified products was visualized and determined in a 1% agarose gel stained with ethidium bromide in 1× Tris-acetate-ethylenediaminetetraacetic acid buffer at 75V for 35–40 min and using a DNA Ladder (ProxiO 1 kb DNA Ladder plus cat. #93406, SRL). The gel images were captured in ultraviolet transillumination with Gel Doc™ XR+ imaging system (Bio-Rad, USA) [Figure 2].
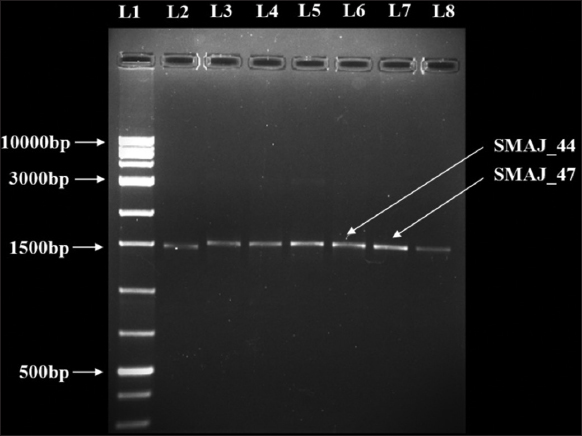 | Figure 2: Agarose gel electrophoresis was performed to analyze the 16S rRNA gene amplified from bacterial isolates. The PCR-amplified products were separated on a 1% agarose gel, and L1 represents the DNA ladder, used as a molecular size reference. [Click here to view] |
2.3.3. Cleanup and purification of the PCR products
PCR products were excised from agarose gels using surgical blades. The gel pieces were kept as small as possible to minimize dilution of the recovered DNA. PCR products were purified using the Wizard® SV Gel and PCR Clean-up System (cat. #A9281, Promega) according to the manufacturer’s instructions [26]. The purity of the extracted DNA samples was assessed by measuring the A260/A280 absorbance ratio, using a Synergy H1 microplate reader (BioTek, USA).
2.3.4. Sequencing and phylogenetic tree construction
The purified PCR products were subsequently sequenced using Sanger sequencing (Eurofins Genomics India Pvt. Ltd). PCR product sequences were assembled using BioEdit software [27]. The partial 16S rDNA sequences of the bacterial strains were analyzed using the basic local alignment search tool (BLAST) algorithm and compared against reference sequences in the NCBI GenBank database for identification. Multiple sequence alignments were conducted using ClustalW via MEGA 11 software [28]. Phylogenetic dendrograms were then constructed using the neighbor-joining (NJ) method based on 16S rDNA sequences to calculate evolutionary distances using MEGA 11 [29,30]. Tree topologies were generated with bootstrap values set at 1000 replicates, to achieve consistent reliability estimates for each clade, with a cut-off point set at 50% for bootstrap replication. The 16S rDNA sequences for bacterial strains SMAJ_44 and SMAJ_47 were submitted to the NCBI GenBank and assigned the accession numbers PP837545 and PP837546, respectively.
2.4. Screening of Bacterial Strains for Abiotic Stress
2.4.1. Determination of HM tolerance
In this study, five HMs, arsenic (As) as (NaAsO2), cadmium (Cd) as (3CdSO4•8H2O), chromium (Cr) as (K2Cr2O7), cobalt (Co) as (CoCl2•6H2O), and copper (Cu) as (CuSO4•5H2O), were used to evaluate bacterial tolerance. The HM salt solutions were prepared by dissolving the respective salts in distilled water, followed by filtration using 0.2 µm pore filters. Each metal was supplemented at varying concentrations (0–1000 µM, while 0–10 mM for As and Cu) in LB broth, pH adjusted to 7.0. A volume of 5 mL medium was inoculated with 50 µL of fresh bacterial suspension equivalent to ~108 CFU/mL. The cultures were then incubated at 28 ± 2°C for 24 h with constant shaking at 100 rpm to assess bacterial growth and tolerance. Followed by measuring the optical density (OD) at 600 nm using a Synergy H1 microplate reader (BioTek, USA). The minimum inhibitory concentration (MIC) was determined based on the lowest concentration at which OD600 measurements indicated a 90% reduction in bacterial growth compared to the control [31]. The inhibition percentage was then determined using the following equation [32,33]. To determine the Cr concentration that causes 50% growth inhibition, the inhibitory concentration (IC50) was calculated based on bacterial growth response [34]. For concentrations where IC50 could not be determined from growth inhibition data, GraphPad Prism software for nonlinear regression analysis was used for analysis, using four-parameter logistic and one-phase binding models [35]. Likewise, MIC and IC50 for HMs, salt, and drought stress were also determined.
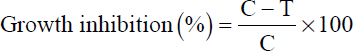 |
Where C is the OD of the control and T is the OD in the presence of HM.
2.4.2. Determination of salt and drought tolerance
The salt tolerance of bacterial strains was evaluated in 5 mL LB broth supplemented with NaCl (0–10% w/v). The cultures were then incubated at 28 ± 2°C for 24 h with constant shaking at 100 rpm to assess bacterial growth and tolerance. Followed by measuring the OD600 [36]. The drought tolerance of bacterial strains was assessed in LB broth supplemented with varying concentrations of PEG 6000 (0–50%) to simulate water stress conditions. The cultures were then incubated at 28 ± 2°C for 24 h with constant shaking at 100 rpm to assess bacterial growth and tolerance. Followed by measuring the OD600 [37].
2.5. PGP Characteristics
2.5.1. Quantitative Assay for Indole Acetic Acid (IAA) Production
Indole acetic acid (IAA) production by strains SMAJ_44 and SMAJ_47 was determined according to Majhi et al. [38] with a few modifications. 5 mL LB broth was inoculated with a fresh culture (~108 CFU/mL) supplemented with 0.1% tryptophan, followed by incubation at 28 ± 2°C for 24 h at 100 rpm. IAA production was quantified by collecting 1.5 mL of culture supernatant and mixing it with 1 mL of Salkowski reagent. The mixture was incubated for 20 min, and absorbance was measured at 530 nm using a spectrophotometer [39,40]. The amount of IAA in the supernatant was quantified using a standard curve created from IAA standard solutions (0–100 µg/mL).
2.5.2. Quantitative assay for siderophore production
For this purpose, SMAJ_44 and SMAJ_47 strains were grown independently in iron-free MM9 medium with modification consisting of (g/L) Sucrose; 10.0, KH2PO4; 1.0, K2HPO4; 0.15, MgSO4•7H2O; 0.2, NH4SO4; 1.0, PIPES; 3.3, NaCl; 5.0, and 1 mL of 100 mM CaCl2 at pH 7.0. For the experiment, 5 mL MM9 medium amended with Cr (VI) (0–1000 µM) after inoculation was incubated for 24 h at 28 ± 2°C, with constant shaking at 100 rpm. For estimation of siderophore, MM9 broth was centrifuged at 10,000 × g for 10 min at 4°C (Eppendorf Centrifuge 5424R) to obtain CFS. The CFS was subjected to a quantitative test using the CAS shuttle assay [41], where 0.5 mL CFS was added to 0.5 mL of CAS reagent, followed by 10 µL of shuttle solution was added and incubated for 30 min. After incubation, absorbance was measured at 630 nm.
 |
Where Ar is the absorbance of the reference (MM9 medium) at 630 nm with CAS reagent, and as is the absorbance of the CFS at 630 nm with CAS reagent.
2.5.3. Quantitative assay for ammonia production
Ammonia production was assessed by growing the bacterial strains in 5 mL of peptone broth supplemented with varying concentrations of Cr (VI) (0–1000 µM) at 28 ± 2°C for 24 h. After incubation, Nessler’s reagent was added to the bacterial suspension, and the development of a yellow to brown coloration was observed as an indicator of ammonia production. The microplate method was used to determine ammonia production by bacterial strains according to Abdelwahed et al. [42]. The absorbance was measured at 450 nm against a standard curve of ammonium sulfate to quantify ammonia production by bacterial strains.
2.5.4. Determination of phosphate (P)-solubilization index
To determine the P-solubilization ability to solubilize sparingly soluble inorganic phosphate, strains were screened for clear phosphate-solubilizing zones around the colonies after 5 days of incubation at 28 ± 2°C on the agar plates with NBRIP (National Botanical Research Institute’s Phosphate) medium consisting of glucose (10 g/L), (NH4)2SO4 (0.1 g/L), MgSO4•7H2O (0.25 g/L), KCl (0.2 g/L), MgCl2•6H2O (5.0 g/L), Ca3(PO4)2 (5 g/L) and agar (15 g/L) [43]. The diameter of the bacterial colony and the halo zone were measured on the 5th day and then were used to evaluate the Phosphate Solubilization Index (PSI) according to the equation below [44].
 |
2.5.5. Production and extraction of EPS
Bacterial strains were cultured in LB broth (50 mL) and incubated at 28 ± 2°C for 24 h. These broth cultures were later centrifuged at 10,000 g for 15 min to separate EPS from bacterial biomass. The EPS was extracted from the cell-free supernatant by adding ice-cold absolute alcohol in the ratio of 1:2 (v/v), and the mixture was stored at 4°C overnight for complete precipitation of EPS [45]. The amount of EPS was determined using the gravimetric method, where the precipitated EPS was collected by centrifugation at 10,000 × g for 10 min to obtain a firm pellet. The supernatant was carefully discarded, and the resulting pellet was air dried for 1 h and weighed using an analytical balance to determine the EPS yield.
2.6. Statistical Analysis
All experiments were conducted in quadruplicate (n = 4), with data presented as means, standard deviation, and standard error. Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) software version 27 for Windows (SPSS Inc., USA.), where an unpaired Student’s t-test was applied to compare differences between two groups, and one-way analysis of variance (ANOVA) was used to assess significant differences among multiple groups. Where a significant difference exists in one-way ANOVA, Tukey’s HSD was used to separate and compare means. Differences in means were considered significant at P ≤ 0.05. Graphs were generated using Microsoft Excel 2013 (Microsoft Corporation, Albuquerque, NM, USA), and IC50 was evaluated using GraphPad Prism 9.3.1.
3. RESULTS AND DISCUSSION
3.1. Morphological and Biochemical Characterization
The morphological characteristics of the bacterial strains SMAJ_44 and SMAJ_47, derived from chickpea root nodules, were comprehensively analyzed in the YMACR plate. Both strains exhibited circular colony shapes with entire margins and raised elevations. The pigmentation differed, with strain SMAJ_44 displaying reddish-pink colonies, which is attributed to the production of prodigiosin, a red-pink secondary metabolite which is typically produced by Serratia spp., as reported in previous scientific studies [46]. Whereas strain SMAJ_47 exhibited peach-pink colored colonies. In terms of optical characteristics, strain SMAJ_44 colonies were opaque, in contrast to the translucent colonies of strain SMAJ_47. Gram staining revealed that both strains were Gram-negative rods arranged singly. Biochemically, both strains were catalase positive. However, a distinction was observed in the coagulase test, where strain SMAJ_44 was coagulase negative, and strain SMAJ_47 was coagulase-positive. Both strains tested negative for the methyl red test and positive for the Voges–Proskauer and citrate test, as shown in Table 1.
Table 1: Morphological and biochemical characteristics of bacterial isolates.
| Morphological and biochemical tests | Bacterial strains | |
|---|---|---|
| SMAJ_44 | SMAJ_47 | |
| Colony shape | Circular | Circular |
| Colony margin | Entire | Entire |
| Elevation | Raised | Raised |
| Pigmentation | Reddish pink | Peach pink |
| Optical characteristics | Opaque | Translucent |
| Gram stain | - | - |
| Cell shape | Rod | Rod |
| Cell arrangement | Single | Single |
| Catalase | + | + |
| Coagulase | − | + |
| Methyl red | − | − |
| Voges proskauer | + | + |
| Citrate | + | + |
+ Positive, −Negative.
3.2. Genetic Characterization
The 16S rRNA gene typically spans approximately 1500 base pairs. Sequencing results indicated that strains SMAJ_44 and SMAJ_47 had nucleotide base sizes ranging from 353 to 394 bp, respectively. The BLAST results in Table 2 show that strain SMAJ_44 has a similarity of 100% with Serratia marcescens AN66. Strain SMAJ_47 had 99.75% similarity with Enterobacter mori IRQBAS47. The NJ method was used to construct the phylogenetic tree. The phylogenetic tree analysis of Serratia spp. SMAJ_44 and Enterobacter spp. SMAJ_47 is shown in Figure 3. The SMAJ_44 strain branched in the clade S. marcescens, whereas the SMAJ_47 strain branched with the clade E. mori. Bacillus cereus, a Gram-positive bacterium, was chosen as the outgroup for the phylogenetic tree.
Table 2: Bacterial strains with their query coverage, top-hit taxonomy (homologous sequence), percent identity, and NCBI accession number.
| Strain | Query coverage (%) | Homology with | Percent identity (%) | Accession |
|---|---|---|---|---|
| SMAJ_44 | 100 | Serratia marcescens AN66 | 100 | PP657487.1 |
| SMAJ_47 | 100 | Enterobacter mori IRQBAS47 | 99.75 | LC427682.1 |
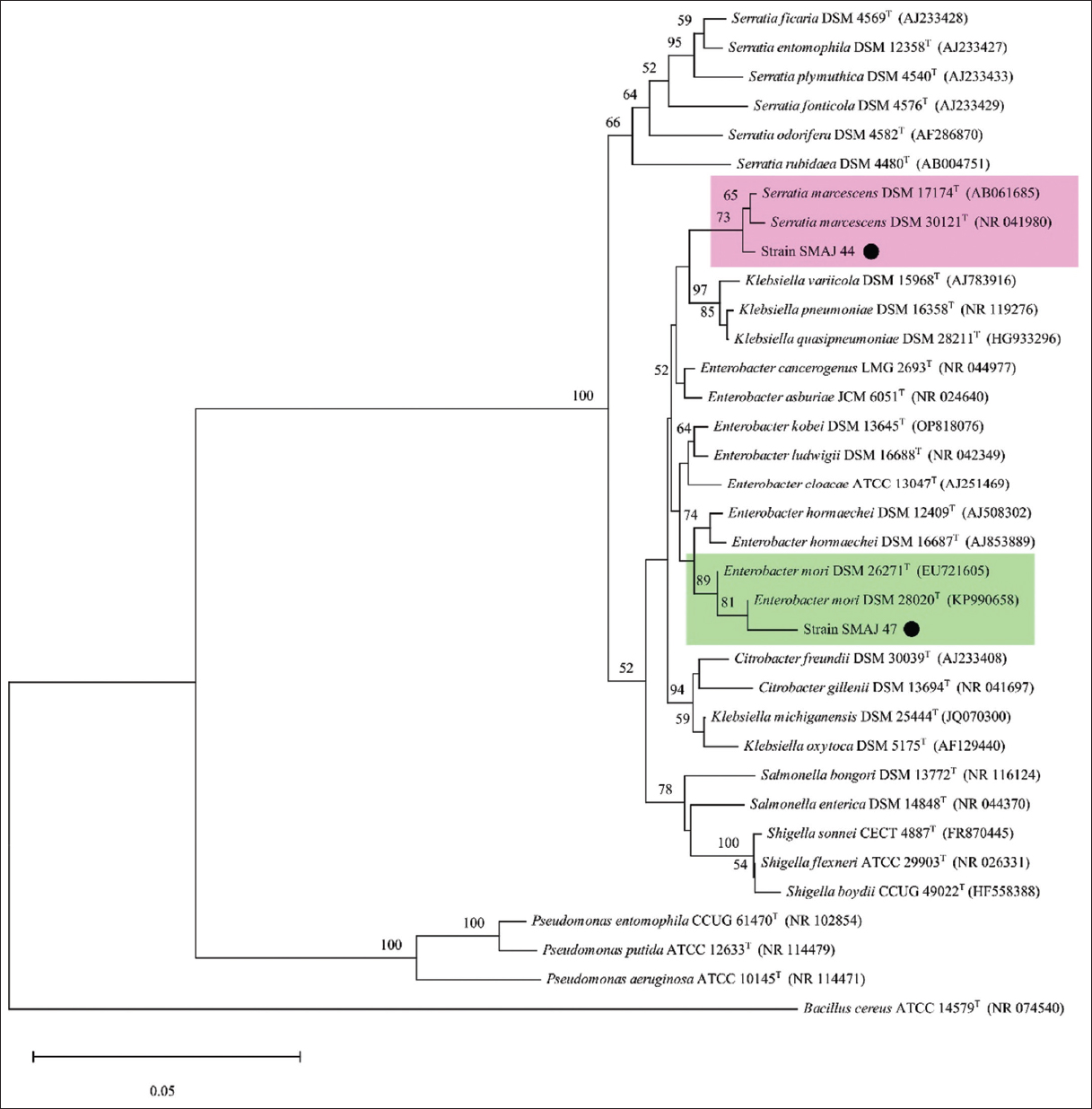 | Figure 3: Phylogenetic tree illustrating the position of strains SMAJ_44 and SMAJ_47, with a black circle (•). The clades for both strains are highlighted. Evolutionary distances were calculated using the maximum composite likelihood, and the topology was inferred using the neighbor-joining method using MEGA 11. Bootstrap values >50% based on 1000 replicates are indicated as percentages at the nodes of the tree. The scale bar represents 0.05 substitutions per nucleotide site. Bacillus cereus was arbitrarily chosen as the outgroup. [Click here to view] |
3.3. Determination of MIC and IC50 Values of Bacterial Strains under Abiotic Stress
Cr (VI) tolerance was evaluated for both the strains SMAJ_44 and SMAJ_47. The effect of Cr (VI) stress was observed for the LB broth. From the results, we can see that in both media, strain SMAJ_44 after 24 h showed higher turbidity compared to strain SMAJ_47 in Figure 4a. Likewise, the growth percentage and OD600 value of all other abiotic stresses for both the strains SMAJ_44 and SMAJ_47 are shown in Figure 4b, Figure 5a and b, Figure 6a and b, Figure 7a and b, Figure 8a and b, Figure 9a and b, and Figure 10a and b. IC50 for both the strains SMAJ_44 and SMAJ_47 in the LB broth were determined to be 500 µM and 270.1 µM, respectively. With increasing Cr (VI) stress, the growth percentage of both strains gradually decreased. The MIC values for Cr (VI) were 700 µM for strain SMAJ_44 and 600 µM for strain SMAJ_47. Similarly, MIC and IC50 for other HMs (As, Cd, Co, and Cu), salt, and drought stress were also determined as shown in Table 3. For metals such as Cd, Cr, and Co, the bacterial tolerance threshold was relatively low (up to ~900 µM), so concentrations were expressed in µM. In contrast, for metals such as As and Cu, the isolates could tolerate much higher concentrations (e.g., 4000–8000 µM). To maintain simplicity, these higher values were converted and represented in mM (e.g., 4 mM, 8 mM). Long-term HM contamination in soil exerts a selective pressure that favors the survival of bacterial species capable of developing resistance to HMs. These bacteria can assist plants in mitigating stress and enhancing growth and productivity by transforming HMs into less toxic forms and altering their availability. A deeper understanding of bacterial resistance mechanisms and their interactions with plants can lead to the development of more effective and targeted bioremediation strategies for HMs in agricultural settings. These resistance mechanisms, which involve complex chemical crosstalk between bacteria and plants, include the production of secondary metabolites and other intricate processes [47]. In this study, we evaluated the IC50 for both strains in the LB broth. As Cr (VI) stress increased, the growth percentage of both strains gradually declined. At 700 and 600 µM, strains SMAJ_44 and SMAJ_47 showed a 90% inhibition [Table 3]. Strain SMAJ_47 exhibited lower IC50 values under all metal stress conditions, including salt and drought stress, indicating lower stress resistance compared to strain SMAJ_44. IC50 of NaCl for strains SMAJ_44 and SMAJ_47 was 5.73 (w/v %) and 4.94 (w/v %), respectively. Razzaghi Komaresofla et al. [48] demonstrated that halotolerant bacteria enhance plant growth under salt stress conditions at 200 mM NaCl. Plants exhibit similar responses to drought and salinity due to water stress. High soil salinity causes osmotic stress, ion toxicity, nutrient imbalance, ROS production, and disrupts photosynthesis. These factors negatively impact plant growth, reducing seed germination, seedling vigor, flowering, and overall yield [49]. IC50 of PEG6000 for strains SMAJ_44 and SMAJ_47 was 10 (w/v %) and 5 (w/v %), respectively. Uzma et al. [50] conducted a study that demonstrated that PGPR with multiple beneficial traits alleviated drought stress and enhanced pod length, number of pods per plant, and seed weight in inoculated plants compared to controls. Similarly, inoculation with drought-tolerant, EPS-producing PGPR led to a 27% and 28% increase in root length in wheat varieties [51].
 | Figure 4: (a) Growth response of strains SMAJ_44 and SMAJ_47 in LB broth under chromium stress, (b) percentage of growth in LB broth under chromium stress. Values are expressed as means ± standard error. Different letters denote significant differences between treatments (P < 0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test. [Click here to view] |
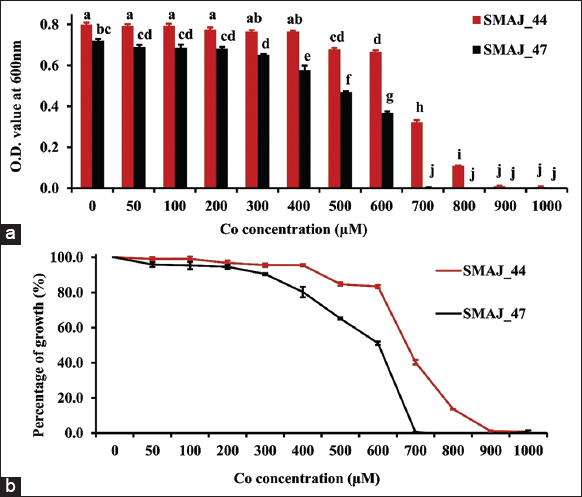 | Figure 5: (a) Growth response of strains SMAJ_44 and SMAJ_47 in LB broth under cobalt stress, (b) Percentage of growth in LB broth under cobalt stress. Values are expressed as means ± standard error. Different letters denote significant differences between treatments (P < 0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test. [Click here to view] |
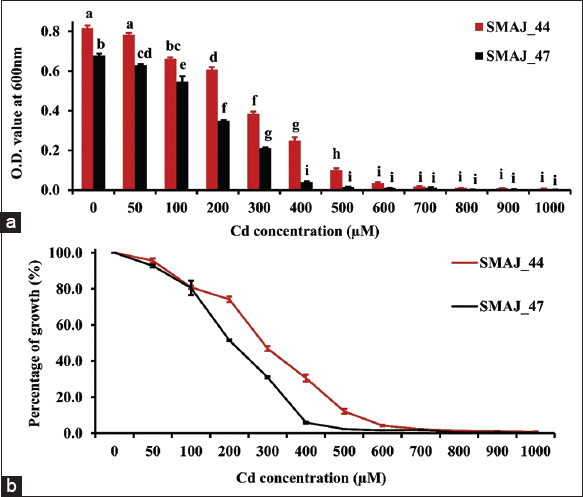 | Figure 6: (a) Growth response of strains SMAJ_44 and SMAJ_47 in LB broth under cadmium stress, (b) percentage of growth in LB broth under cadmium stress. Values are expressed as means ± standard error. Different letters denote significant differences between treatments (P < 0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test. [Click here to view] |
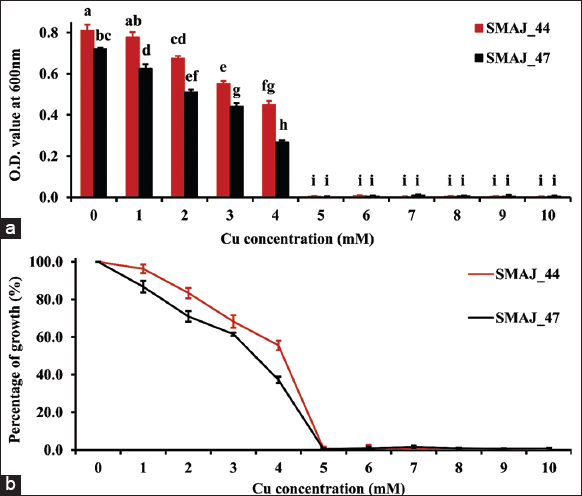 | Figure 7: (a) Growth response of strains SMAJ_44 and SMAJ_47 in LB broth under copper stress, (b) Percentage of growth in LB broth under copper stress. Values are expressed as means ± standard error. Different letters denote significant differences between treatments (P < 0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test. [Click here to view] |
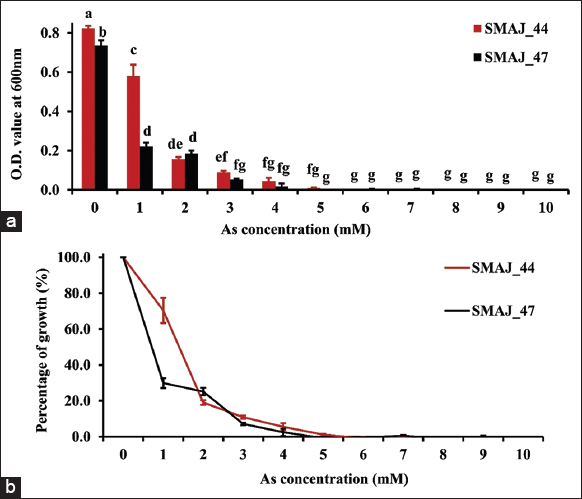 | Figure 8: (a) Growth response of strains SMAJ_44 and SMAJ_47 in LB broth under arsenic stress, (b) Percentage of growth in LB broth under arsenic stress. Values are expressed as means ± standard error. Different letters denote significant differences between treatments (P < 0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test. [Click here to view] |
 | Figure 9: (a) Growth response of strains SMAJ_44 and SMAJ_47 in LB broth under NaCl stress, (b) Percentage of growth in LB broth under NaCl stress. Values are expressed as means ± standard error. Different letters denote significant differences between treatments (P < 0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test. [Click here to view] |
 | Figure 10: (a) Growth response of strains SMAJ_44 and SMAJ_47 in LB broth under PEG6000 stress, (b) Percentage of growth in LB broth under PEG6000 stress. Values are expressed as means ± standard error. Different letters denote significant differences between treatments (P < 0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test. [Click here to view] |
Table 3: Minimum inhibitory concentration (MIC) and IC50 values of bacterial strains under various abiotic stresses.
| Abiotic stress | Strain | Inhibition % | Concentration | Estimated IC50 | MIC |
|---|---|---|---|---|---|
| Cr | SMAJ_44 | 57.1 | 500 µM | ~500 µM | 700 µM |
| SMAJ_47 | 73.03 | 300 µM | ~270.1 µM | 600 µM | |
| Co | SMAJ_44 | 59.66 | 700 µM | ~686.6 µM | 900 µM |
| SMAJ_47 | 53.97 | 600 µM | ~600 µM | 700 µM | |
| Cd | SMAJ_44 | 53.06 | 300 µM | ~300 µM | 600 µM |
| SMAJ_47 | 57.30 | 200 µM | ~200 µM | 400 µM | |
| Cu | SMAJ_44 | 44.43 | 4 mM | ~4.11 mM | 5 mM |
| SMAJ_47 | 62.72 | 4 mM | ~3.48 mM | 5 mM | |
| As | SMAJ_44 | 80.97 | 2 mM | ~1.33 mM | 4 mM |
| SMAJ_47 | 73.26 | 1 mM | ~0.65 mM | 3 mM | |
| NaCl | SMAJ_44 | 60.23 | 6 (w/v %) | ~5.73 (w/v %) | 8 (w/v %) |
| SMAJ_47 | 85.82 | 6 (w/v %) | ~4.94 (w/v %) | 8 (w/v %) | |
| PEG6000 | SMAJ_44 | 53.95 | 10 (w/v %) | ~10 (w/v %) | 30 (w/v %) |
| SMAJ_47 | 53.25 | 5 (w/v %) | ~5 (w/v %) | 20 (w/v %) |
~Approximately.
3.4. Quantification of PGP Traits
3.4.1. IAA production
The PGP traits of bacterial strains SMAJ_44 and SMAJ_47 were assessed based on their ability to produce IAA, ammonia, siderophores, phosphate solubilization, and EPS production. The results revealed that strain SMAJ_47 exhibited significantly higher IAA production, 1037.69 ± 21.50 (P < 0.001), in comparison to strain SMAJ_44, which exhibited 90.78 ± 5.68 µg/mL production, suggesting its greater potential in promoting root elongation and plant development. In the treatment set for strain SMAJ_47, IAA production at 50µM Cr (VI) showed a significant increase of 1135.94 ± 15.68 (P < 0.001) compared to the control 1037.69 ± 21.50 [Table 4]. However, in strain SMAJ_44, IAA levels at 0µM and 50µM Cr (VI) were comparable (P > 0.05), indicating no significant change in response to the Cr (VI) stress. IAA is a phytohormone produced by plants and various microorganisms. It enhances plant growth and plays a crucial role in the interaction between plants and microorganisms [52]. IAA production by bacteria can vary significantly across different species and strains, and it is influenced by factors such as culture conditions, substrate availability, and growth stage [53]. The experimental results demonstrated that SMAJ_44 and SMAJ_47 strains successfully utilized L-tryptophan as a precursor to produce IAA under in vitro conditions. Rizvi and Khan [54] reported similar findings that IAA production by Pseudomonas aeruginosa decreased with increasing concentrations of Cd, Cr, and Cu, ranging from 25 to 400 µg/mL. IAA produced by PGPB enhances plant growth and colonization. It was reported that bacterial inoculation improved plant growth and physiology, resulting in better survival rates, increased root/shoot biomass, and higher nutrient content [55]. IAA-producing bacteria promote plant growth, as reported by Shahzad et al. [56], demonstrated endophytic bacteria significantly increased root and shoot length, fresh and dry weight of the plant, and chlorophyll content of Oryza sativa compared to the control group treated with distilled water and E. coli, which did not produce IAA.
Table 4: Plant growth-promoting (PGP) traits of bacterial strains.
| PGP traits | Cr (VI) (µM) | Strain SMAJ_44 | Strain SMAJ_47 |
|---|---|---|---|
| IAA production (µg/mL) | 0 | 90.78±5.68d | 1037.69±21.50b |
| 50 | 82.28±1.68d | 1135.94±15.68a | |
| 100 | 82.05±1.16d | 289.34±8.40c | |
| 200–1000 | - | - | |
| Ammonia production (mM) | 0 | 12.38±0.99ab | 11.17±0.53abc |
| 50 | 12.90±1.85a | 10.59±0.65bc | |
| 100 | 11.68±0.62ab | 5.41±0.58d | |
| 200 | 9.41±0.36c | - | |
| 300–1000 | - | - | |
| Siderophore production (%) | 0 | 45.33±1.08a | 37.00±2.23b |
| 50 | 32.99±2.42b | 24.73±1.84c | |
| 100 | 11.93±3.44d | 8.00±0.73d | |
| 200–1000 | - | - |
Values represent means of replicates (n=4); mean values (mean±standard deviation); -, No production. Different letters denote significant differences between treatments (P<0.05) according to One-way analysis of variance with Tukey’s post hoc multiple comparison test.
3.4.2. Siderophore production
Siderophore production was higher in strain SMAJ_44 (45.33 ± 1.08 SU) (P < 0.05) than in strain SMAJ_47 (37.00 ± 2.23 SU), suggesting a stronger ability to enhance iron availability for plants. At 50µM Cr (VI), strain SMAJ_47 exhibited a significant decrease (P < 0.001) in siderophore production, whereas strain SMAJ_44 maintained production levels comparable to the control. A similar pattern of tolerance at lower Cr (VI) concentrations was also observed in IAA production, where strain SMAJ_44 showed stable output despite the Cr (VI) stress. Since iron is often poorly soluble in soil, both plants and microorganisms utilize distinct strategies, such as siderophore secretion, to increase iron solubility and facilitate uptake. Given that both strains demonstrated siderophore production, they hold promise for use as bioinoculants with potential biotechnological applications in agriculture, particularly in enhancing crop growth and nutrient acquisition under iron-limited conditions. It is important to note that bacterial growth and siderophore production are influenced by various factors, including the type of carbon source (sucrose, fructose, glucose, glycerol, mannose, maltose, and xylose) used. Moreover, concentrations of metal (Fe3+, Mn2+, Cu2+, and Zn2+), different nitrogen sources (asparagine, glutamic, lysine, proline, tyrosine, and tryptophan), pH, temperature, and incubation time [57]. This study used sucrose as a carbon source to estimate siderophore production in MM9 medium. In our study, siderophore production decreased as Cr (VI) stress levels increased. Similar findings were reported by Shi et al. [58], who demonstrated that at high concentrations (1000 µM) of HMs (Cd2+, Cu2+, Mn2+, Ni2+, Pb2+, and Zn2+), total siderophore production in P. aeruginosa decreased. Conversely, at lower concentrations (200–400 µM), siderophore production levels were elevated. Notably, Cu2+ significantly inhibited the total siderophore and pyoverdine production in P. aeruginosa. According to Braud et al. [59], siderophores such as pyoverdine and pyochelin can sequester metals from the extracellular environment of bacteria, thereby reducing metal diffusion into the cells. This indicates that siderophores may be produced not only for iron acquisition but also to sequester toxic metals under certain conditions. In our study, siderophore production was significantly inhibited at Cr (VI) concentrations exceeding 200µM [Table 4].
3.4.3. Ammonia production
In terms of ammonia production, no statistically significant difference was observed (P > 0.05) in the control set between strain SMAJ_44 and SMAJ_47. Both strains showed similar capabilities, with strain SMAJ_44 producing 12.38 ± 0.99 mM and strain SMAJ_47 producing 11.17 ± 0.53 mM [Table 4], indicating their role in nitrogen enrichment of the soil. However, above 300 µM, the production of ammonia was not detected for both strains. These findings indicate a threshold beyond which Cr (VI) toxicity adversely affects the functional capabilities of beneficial bacteria, limiting their role in alleviating HM stress for plants.
3.4.4. P-solubilization index
Phosphate solubilization, a crucial factor for improving phosphorus uptake, was slightly more efficient in strain SMAJ_47 showed a solubilization index of 2.73 ± 0.27 (P < 0.05) than in strain SMAJ_44 (3.38 ± 0.28), as shown in Table 5, highlighting its better potential in mobilizing phosphorus [Figure 11]. Phosphate-solubilizing bacteria (PSB) provide a promising approach to enhance phosphorus availability in the soil. Several studies have demonstrated the PGP effects of PSB on maize, chickpea, barley, lettuce, and other plants [60]. These bacteria serve as a biotechnological alternative to chemical fertilizers.
Table 5: Phosphate solubilization index (PSI) and exopolysaccharide (EPS) production by bacterial strains.
| Bacterial strains | PSI | EPS (g/L) |
|---|---|---|
| SMAJ_44 | 2.73±0.27a | 0.60±0.22a |
| SMAJ_47 | 3.38±0.28b | 0.64±0.76a |
Values represent means of replicates (n=4); mean values (mean±standard deviation). Different letters denote significant differences between treatments (P<0.05), data analyzed using the unpaired Student’s t-test.
 | Figure 11: Phosphate-solubilizing halo zones formed by bacterial strains SMAJ_44 and SMAJ_47 on NBRIP agar plates. Sterile LB broth was used as a control. [Click here to view] |
Overall, these findings suggest that both strains exhibit promising PGP traits, with strain SMAJ_47 excelling in IAA production and phosphate solubilization, whereas strain SMAJ_44 demonstrates superior siderophore production, making them potential candidates for improving plant growth under nutrient-deficient conditions. At concentrations exceeding 300 µM of Cr (VI), PGP production by both bacterial strains was significantly inhibited. This reveals that higher Cr levels cause significant stress on bacterial metabolism, potentially disrupting key pathways involved in ammonia production, IAA production, and siderophore secretion. Thus, higher levels of Cr may lead to oxidative stress, reduced nutrient uptake efficiency, and overall metabolic suppression, thereby reducing the ability of bacteria to facilitate plant growth under higher metal-contaminated conditions.
3.4.5. Production of EPS
PGPB produces EPS and biosurfactants that maintain soil structure and fertility, even under HM stress. The polysaccharides and lipopolysaccharides in EPS facilitate the removal of metals from rhizospheric soil through biosorption, which is mediated by anionic functional groups such as phosphate, hydroxyl, succinyl, and uronic acids. These functional groups enable EPS to mobilize toxic metals effectively [61]. Serratia spp. and Enterobacter spp. have both been previously reported to produce EPS [62-67]. Our findings indicate that there was no statistically significant difference (P > 0.05) in EPS production between the two bacterial strains [Table 5], with strain SMAJ_44 producing 0.60 ± 0.22 g/L and strain SMAJ_47 producing 0.64 ± 0.76 g/L [Figure 12], which is beneficial for biofilm formation and stress tolerance. These findings suggest our bacterial strains can play an important role in sustainable agriculture by enhancing soil health, elevating plant stress tolerance, and aiding in bioremediation. All these PGP attributes make them a promising tool for developing sustainable agriculture systems.
 | Figure 12: Exopolysaccharides production by bacterial strains SMAJ_44 and SMAJ_47 was obtained from 25 mL LB broth. [Click here to view] |
4. CONCLUSION
The two bacterial strains isolated from chickpea root nodules were evaluated for their tolerance to HMs including chromium (Cr), cadmium (Cd), copper (Cu), cobalt (Co), and arsenic (As), with particular emphasis on Cr (VI), as it served as the primary focus of this study, under in vitro conditions. The two strains isolated from chickpea root nodules demonstrated tolerance to HMs and possessed PGP traits, which may enhance the growth and survival of the host plant in a nutrient-deficient and abiotic stress environment. Strain SMAJ_44 exhibited higher siderophore production, whereas strain SMAJ_47 produced higher IAA. However, under higher Cr (VI) stress, all PGP traits, such as IAA, ammonia, and siderophore production, were not detected. Along with salinity and drought stress, HM tolerance is crucial for PGPR to mitigate HM toxicity and preserve their PGP traits under HM stress conditions. Utilizing abiotic stress-tolerant bacteria that produce PGP traits, as bioinoculants, is highly advantageous for boosting crop yield and preserving soil fertility in both agricultural fields and tree-based ecosystems in saline, drought, and HM-contaminated lands for sustainable agriculture. Recent advancements in microbial biotechnology and bacterial transformation systems may facilitate the engineering of endophytic bacteria to enhance crop benefits. The performance of the inoculant could be improved greatly if it could have higher resistance against HM stress and abundantly express one or more PGP traits that interact synergistically.
5. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be authors as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
6. ACKNOWLEDGMENT
The graphical abstract was generated using Inkscape software (https://inkscape.org/) and Bioicons (https://bioicons.com/). The authors acknowledge Prerona Biswas, Sima Sikdar, and Aparna Shil for critically reading the manuscript and for their valuable input. We also acknowledge the infrastructural support received from Presidency University, Kolkata, for preparing the manuscript.
7. FUNDING
The authors gratefully acknowledge the financial support provided by the FRPDF (Faculty Research and Professional Development Fund) Grant of Presidency University, Kolkata, to the corresponding author, the DST-FIST Program of the Department of Life Sciences, Presidency University, Kolkata [Grant no. SR/FST/LSI-560/2013(c); dated June 23, 2015] and the DBT-BUILDER Grant of the Department of Life Sciences, Presidency University, Kolkata [Grant no. BT/INF/22/SP45088/2022; dated February 17, 2022]. University Grants Commission National Eligibility Test Fellowship Grant [F.16-6(DEC. 2016)/2017(NET), UGC-Ref. No.: 764/(ST)(CSIR-UGC NET DEC. 2016)] to Mr. Sudipta Majhi.
8. CONFLICTS OF INTEREST
The authors declare that they have no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data are available to the authors and shall be provided upon request. The 16S rDNA sequences of the bacterial strains have been submitted to the National Centre for Biotechnology Information (https://ncbi.nlm.nih.gov). Genebank database under the accession numbers PP837545 (S. marcescens SMAJ_44) and PP837546 (E. mori SMAJ_47) are available online in the NCBI gene database.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declare that artificial intelligence (AI)-assisted tools were used solely for improving the readability of the manuscript, including grammar, spelling, and language refinement. No AI tools were employed for result generation, data analysis, or image manipulation.
REFERENCES
1. Mishra S, Bharagava RN, More N, Yadav A, Zainith S, Mani S, et al. Heavy metal contamination:An alarming threat to environment and human health. In: Environmental Biotechnology: For Sustainable Future. Singapore: Springer Singapore;2019. 103-25.[CrossRef]
2. Yang Y, Ding J, Chi Y, Yuan J. Characterization of bacterial communities associated with the exotic and heavy metal tolerant wetland plant Spartina alterniflora. Sci Rep. 2020;10:17985.[CrossRef]
3. El-Ballat EM, Elsilk SE, Ali HM, Ali HE, Hano C, El-Esawi MA. Metal-resistant PGPR strain Azospirillum brasilense EMCC1454 enhances growth and chromium stress tolerance of chickpea (Cicer arietinum L.) by modulating redox potential, osmolytes, antioxidants, and stress-related gene expression. Plants. 2023;12:2110.[CrossRef]
4. Hossini H, Shafie B, Niri AD, Nazari M, Esfahlan AJ, Ahmadpour M, et al. A comprehensive review on human health effects of chromium:Insights on induced toxicity. Environ Sci Pollut Res. 2022;29:70686-705.[CrossRef]
5. International Agency for Research on Cancer. Agents Classified by the IARC Monographs. IGARSS 2014. France:International Agency for Research on Cancer;2012.
6. Majhi S, Sikdar M. How heavy metal stress affects the growth and development of pulse crops:Insights into germination and physiological processes. 3 Biotech. 2023;13:155.[CrossRef]
7. Tirry N, Kouchou A, El Omari B, Ferioun M, El Ghachtouli N. Improved chromium tolerance of Medicago sativa by plant growth-promoting rhizobacteria (PGPR). J Genet Eng Biotechnol. 2021;19:149.[CrossRef]
8. Ajmal AW, Saroosh S, Mulk S, Hassan MN, Yasmin H, Jabeen Z, et al. Bacteria isolated from wastewater irrigated agricultural soils adapt to heavy metal toxicity while maintaining their plant growth promoting traits. Sustainability. 2021;13:7792.[CrossRef]
9. Kong Z, Wu Z, Glick BR, He S, Huang C, Wu L. Co-occurrence patterns of microbial communities affected by inoculants of plant growth-promoting bacteria during phytoremediation of heavy metal-contaminated soils. Ecotoxicol Environ Saf. 2019;183:109504.[CrossRef]
10. Ullah A, Heng S, Munis MF, Fahad S, Yang X. Phytoremediation of heavy metals assisted by plant growth promoting (PGP) bacteria:A review. Environ Exp Bot. 2015;117:28-40.[CrossRef]
11. Akhtar N, Ilyas N, Mashwani ZR, Hayat R, Yasmin H, Noureldeen A, et al. Synergistic effects of plant growth promoting rhizobacteria and silicon dioxide nano-particles for amelioration of drought stress in wheat. Plant Physiol Biochem. 2021;166:160-76.[CrossRef]
12. Riskuwa-Shehu ML, Ismail HY, Ijah UJ. Heavy metal resistance by endophytic bacteria isolated from guava (Psidium Guajava) and mango (Mangifera Indica) Leaves. Int Ann Sci. 2019;9:16-23. https://doi.org/10.21467/ias.9.1.16-23 [CrossRef]
13. Zahra ST, Tariq M, Abdullah M, Ullah MK, Rafiq AR, Siddique A, et al. Salt-tolerant plant growth-promoting bacteria (ST-PGPB):An effective strategy for sustainable food production. Curr Microbiol. 2024;81:304.[CrossRef]
14. Khan N, Bano A, Rahman MA, Guo J, Kang Z, Babar MA. Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci Rep. 2019;9:2097.[CrossRef]
15. Kalsoom A, Batool R, Jamil N. Beneficial rhizospheric associated traits of chromate resistant bacteria for remediation of Cr (VI) contaminated soil. Bioremediat J. 2023;27:189-207.[CrossRef]
16. Kumar A, Tripti, Maleva M, Bruno LB, Rajkumar M. Synergistic effect of ACC deaminase producing Pseudomonas sp. TR15a and siderophore producing Bacillus aerophilus TR15c for enhanced growth and copper accumulation in Helianthus annuus L. Chemosphere. 2021;276:130038.[CrossRef]
17. Basu A, Prasad P, Das SN, Kalam S, Sayyed RZ, Reddy MS, et al. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants:Recent developments, constraints, and prospects. Sustainability. 2021;13:1140. [CrossRef]
18. Misra S, Semwal P, Pandey DD, Mishra SK, Chauhan PS. Siderophore-producing Spinacia oleracea bacterial endophytes enhance nutrient status and vegetative growth under iron-deficit conditions. J Plant Growth Regul. 2024;43:1317-30.[CrossRef]
19. Blaga AC, Zaharia C, Suteu D. Polysaccharides as support for microbial biomass-based adsorbents with applications in removal of heavy metals and dyes. Polymers (Basel). 2021;13:2893.[CrossRef]
20. Thai TD, Lim W, Na D. Synthetic bacteria for the detection and bioremediation of heavy metals. Front Bioeng Biotechnol. 2023;11:1-21.[CrossRef]
21. Liu Y, Li J, Du J, Hu M, Bai H, Qi J, et al. Accurate assessment of antibiotic susceptibility and screening resistant strains of a bacterial population by linear gradient plate. Sci China Life Sci. 2011;54:953-960.[CrossRef]
22. Holt JG, Krieg NR, Sneath PH, Staley JT, Williams ST. Bergey's Manual of Determinative Bacteriology. 9th ed. Baltimore:Lippincott Williams and Wilkins;2000.
23. Anumalla M, Batchu UR, Singh NK, Bhukya B, Reddy Shetty P. Draft genome analysis of Delftia tsuruhatensis IICT-RSP4, a strain with uricase potential isolated from soil. Indian J Microbiol. 2025.[CrossRef]
24. Li J, Liu P, Menguy N, Benzerara K, Bai J, Zhao X, et al. Identification of sulfate-reducing magnetotactic bacteria via a group-specific 16S rDNA primer and correlative fluorescence and electron microscopy:Strategy for culture-independent study. Environ Microbiol. 2022;24:5019-38.[CrossRef]
25. Mukherjee S, De A, Sarkar NK, Saha NC. Isolation and characterization of benzene utilizing Bacillus spp. from petroleum contaminated soil in Kolkata, West Bengal, India. Asian J Microbiol Biotechnol Environ Sci. 2019;21:428-37.
26. Kabir MM, Fakhruddin AN, Chowdhury MA, Pramanik MK, Fardous Z. Isolation and characterization of chromium(VI)-reducing bacteria from tannery effluents and solid wastes. World J Microbiol Biotechnol. 2018;34:126. [CrossRef]
27. Mekonnen E, Kebede A, Nigussie A, Kebede G, Tafesse M. Isolation and characterization of urease-producing soil bacteria. Int J Microbiol. 2021;2021:1-11.[CrossRef]
28. Sadiqi S, Hamza M, Ali F, Alam S, Shakeela Q, Ahmed S, et al. Molecular characterization of bacterial isolates from soil samples and evaluation of their antibacterial potential against MDRS. Molecules. 2022;27:6281. [CrossRef]
29. Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. 2004;101:11030-5.[CrossRef]
30. Tamura K, Stecher G, Kumar S. MEGA11:Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022-7.[CrossRef]
31. Xu Y, Tan L, Li Q, Zheng X, Liu W. Sublethal concentrations of heavy metals Cu2+ and Zn2+ can induce the emergence of bacterial multidrug resistance. Environ Technol Innov. 2022;27:102379.[CrossRef]
32. Firinc?C, Zamfir LG, Constantin M, R?ut I, Capr?L, Popa D, et al. Microbial removal of heavy metals from contaminated environments using metal-resistant indigenous strains. J Xenobiot. 2023;14:51-78.[CrossRef]
33. Yaghoubian Y, Siadat SA, Moradi Telavat MR, Pirdashti H, Yaghoubian I. Bio-removal of cadmium from aqueous solutions by filamentous fungi:Trichoderma spp. and Piriformospora indica. Environ Sci Pollut Res. 2019;26:7863-72.[CrossRef]
34. Zutshi S, Choudhary M, Bharat N, Abdin MZ, Fatma T. Evaluation of antioxidant defense responses to lead stress in Hapalosiphon fontinalis ?339(1). J Phycol. 2008;44:889-96.[CrossRef]
35. Shukor MY, Masdor N, Baharom NA, Jamal JA, Abdullah MP, Shamaan NA, et al. An inhibitive determination method for heavy metals using bromelain, a cysteine protease. Appl Biochem Biotechnol. 2008;144:283-91. [CrossRef]
36. Afridi MS, Van Hamme JD, Bundschuh J, Sumaira, Khan MN, Salam A, et al. Biotechnological approaches in agriculture and environmental management - bacterium Kocuria rhizophila 14ASP as heavy metal and salt- tolerant plant growth- promoting strain. Biologia (Bratisl). 2021;76:3091-105.[CrossRef]
37. Agunbiade VF, Fadiji AE, Agbodjato NA, Babalola OO. Isolation and characterization of plant-growth-promoting, drought-tolerant rhizobacteria for improved maize productivity. Plants. 2024;13:1298.[CrossRef]
38. Majhi K, Let M, Bandopadhyay R. Efficacious use of Micrococcus yunnanensis GKSM13 for the growth of rice seedlings under copper stress with elucidation into genomic traits. Curr Plant Biol. 2024;37:100318.[CrossRef]
39. Gordon SA, Weber RP. Colorimetric estimation of indoleacetic acid. Plant Physiol. 1951;26:192-5.[CrossRef]
40. Pinto MS, Inocente LB, Oliveira PN, Silva KJ, Carrer H. Plant growth-promoting (PGP) traits of endophytic bacteria from in vitro cultivated Tectona grandis L.f. Forests. 2022;13:1539.[CrossRef]
41. Payne SM. Detection, isolation, and characterization of siderophores. Methods Enzymol. 1994;235:329-44.[CrossRef]
42. Abdelwahed S, Trabelsi E, Saadouli I, Kouidhi S, Masmoudi AS, Cherif A, et al. A new pioneer colorimetric micro-plate method for the estimation of ammonia production by plant growth promoting rhizobacteria (PGPR). Main Gr Chem. 2022;21:55-68.[CrossRef]
43. Kshetri L, Pandey P, Sharma GD. Rhizosphere mediated nutrient management in Allium hookeri Thwaites by using phosphate solubilizing rhizobacteria and tricalcium phosphate amended soil. J Plant Interact. 2018;13:256-69.[CrossRef]
44. Doilom M, Guo JW, Phookamsak R, Mortimer PE, Karunarathna SC, Dong W, et al. Screening of phosphate-solubilizing fungi from air and soil in Yunnan, China:Four novel species in Aspergillus, Gongronella, Penicillium, and Talaromyces. Front Microbiol. 2020;11:585215.[CrossRef]
45. Sathishkumar R, Kannan R, Jinendiran S, Sivakumar N, Selvakumar G, Shyamkumar R. Production and characterization of exopolysaccharide from the sponge-associated Bacillus subtilis MKU SERB2 and its in-vitro biological properties. Int J Biol Macromol. 2021;166:1471-9.[CrossRef]
46. Rodríguez J, Lobato C, Vázquez L, Mayo B, Flórez AB. Prodigiosin-producing Serratia marcescens as the causal agent of a red colour defect in a blue cheese. Foods. 2023;12:2388.[CrossRef]
47. Barra Caracciolo A, Terenzi V. Rhizosphere microbial communities and heavy metals. Microorganisms. 2021;9:1462.[CrossRef]
48. Razzaghi Komaresofla B, Alikhani HA, Etesami H, Khoshkholgh-Sima NA. Improved growth and salinity tolerance of the halophyte Salicornia sp. by co-inoculation with endophytic and rhizosphere bacteria. Appl Soil Ecol. 2019;138 160-170.[CrossRef]
49. Gamalero E, Favale N, Bona E, Novello G, Cesaro P, Massa N, et al. Screening of bacterial endophytes able to promote plant growth and increase salinity tolerance. Appl Sci. 2020;10:5767.[CrossRef]
50. Uzma M, Iqbal A, Hasnain S. Drought tolerance induction and growth promotion by indole acetic acid producing Pseudomonas aeruginosa in Vigna radiata. PLoS One. 2022;17:e0262932.[CrossRef]
51. Latif M, Bukhari SA, Alrajhi AA, Alotaibi FS, Ahmad M, Shahzad AN, et al. Inducing drought tolerance in wheat through exopolysaccharide-producing rhizobacteria. Agronomy. 2022;12:1140.[CrossRef]
52. ALKahtani MD, Fouda A, Attia KA, Al-Otaibi F, Eid AM, Ewais EE, et al. Isolation and characterization of plant growth promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy. 2020;10:1325.[CrossRef]
53. Khiangte L, Lalfakzuala R. Effects of heavy metals on phosphatase enzyme activity and indole-3-acetic acid (IAA) production of phosphate solubilizing bacteria. Geomicrobiol J. 2021;38:494-503.[CrossRef]
54. Rizvi A, Khan MS. Biotoxic impact of heavy metals on growth, oxidative stress and morphological changes in root structure of wheat (Triticum aestivum L.) and stress alleviation by Pseudomonas aeruginosa strain CPSB1. Chemosphere. 2017;185:942-52.[CrossRef]
55. Naveed M, Qureshi MA, Zahir ZA, Hussain MB, Sessitsch A, Mitter B. L-Tryptophan-dependent biosynthesis of indole-3-acetic acid (IAA) improves plant growth and colonization of maize by Burkholderia phytofirmans PsJN. Ann Microbiol. 2015;65:1381-9.[CrossRef]
56. Shahzad R, Waqas M, Khan AL, Al-Hosni K, Kang SM, Seo CW, et al. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol Hung. 2017;68:175-86.[CrossRef]
57. Sheng MM, Jia HK, Zhang GY, Zeng LN, Zhang TT, Long YH, et al. Siderophore production by rhizosphere biological control bacteria Brevibacillus brevis GZDF3 of Pinellia ternata and its antifungal effects on Candida albicans. J Microbiol Biotechnol. 2020;30:689-99.[CrossRef]
58. Shi P, Xing Z, Zhang Y, Chai T. Effect of heavy-metal on synthesis of siderophores by Pseudomonas aeruginosa ZGKD3. IOP Conf Ser Earth Environ Sci. 2017;52:012103.[CrossRef]
59. Braud A, Geoffroy V, Hoegy F, Mislin GL, Schalk IJ. Presence of the siderophores pyoverdine and pyochelin in the extracellular medium reduces toxic metal accumulation in Pseudomonas aeruginosa and increases bacterial metal tolerance. Environ Microbiol Rep. 2010;2:419-25.[CrossRef]
60. Lucero CT, Lorda GS, Anzuay MS, Ludueña LM, Taurian T. Peanut endophytic phosphate solubilizing bacteria increase growth and P content of soybean and maize plants. Curr Microbiol. 2021;78:1961-72.[CrossRef]
61. Bhagat N, Raghav M, Dubey S, Bedi N. Bacterial exopolysaccharides:Insight into their role in plant abiotic stress tolerance. J Microbiol Biotechnol. 2021;31:1045-59.[CrossRef]
62. Almutairi MH, Helal MM. Exopolysaccharide production from isolated Enterobacter sp. strain ACD2 from the northwest of Saudi Arabia. J King Saud Univ Sci. 2021;33:101318.[CrossRef]
63. Prakash Shyam K, Rajkumar P, Ramya V, Sivabalan S, Kings AJ, et al. Exopolysaccharide production by optimized medium using novel marine Enterobacter cloacae MBB8 isolate and its antioxidant potential. Carbohydr Polym Technol Appl. 2021;2:100070.[CrossRef]
64. Bezawada J, Hoang NV, More TT, Yan S, Tyagi N, Tyagi RD, et al. Production of extracellular polymeric substances (EPS) by Serratia sp.1 using wastewater sludge as raw material and flocculation activity of the EPS produced. J Environ Manage. 2013;128:83-91.[CrossRef]
65. Khan A, Singh AV. Multifarious effect of ACC deaminase and EPS producing Pseudomonas sp. and Serratia marcescens to augment drought stress tolerance and nutrient status of wheat. World J Microbiol Biotechnol. 2021;37:198.[CrossRef]
66. Leng F, Zhang K, Hu S, Li S, Yu C, Wang Y. Exopolysaccharides of Serratia fonticola CPSE11 can alleviate the toxic effect of Cd2+ on Codonopsis pilosula. Environ Sci Pollut Res. 2023;30:80378-92.[CrossRef]
67. Dhanya BE, Athmika, Rekha PD. Characterization of an exopolysaccharide produced by Enterobacter sp. YU16-RN5 and its potential to alleviate cadmium induced cytotoxicity in vitro. 3 Biotech. 2021;11:491.[CrossRef]