1. INTRODUCTION
Vibrio parahaemolyticus has been one of the causes of acute hepatopancreatic necrosis disease or early mortality syndrome in shrimps in recent years. Using antibiotics or chemicals to inhibit bacteria has been the most popular course of treatment due to the initial benefits they bring. However, the long-term use of antibiotics leads to antibiotic resistance in bacteria, which is still a challenge in the aquaculture industry and the field of shrimp farming. The research on the inhibition of V. parahaemolyticus using biological therapy is being considered. The method of using phages against aquatic pathogenic bacteria was first used in Japan [1] and quickly became the subject of scientific interest. In recent years, there have been a number of studies using phages in the prevention and treatment of plant and animal diseases [2-4]. Besides, there are also many studies on plant extracts with high antimicrobial efficiency [5], especially Phyllanthus amarus [6,7]. Bacteriophages and P. amarus are considered potential biological agents in the treatment of bacterial diseases in aquatic animals. At present, research and potential applications of these agents in practice are underway, but studies in our country that involve their combined use are still limited. Therefore, the study of the ability of the phage in combination with the extract of this plant to inhibit the V. parahaemolyticus bacteria serves as a scientific basis for herbal applications in aquaculture in general and shrimp farming. The objective of this study was to investigate the interaction of phage and P. amarus extract and their effectiveness in inhibiting the spread of the pathogenic V. parahaemolyticus on white leg shrimp.
2. MATERIALS AND METHODS
2.1. Materials
All chemicals, equipment, and bacteriophages were supplied by the Molecular Biology Laboratory, Biotechnology Research and Development Institute, Cantho University. The methanolic extract of P. amarus (all the body) was obtained by the College of Natural Sciences, Cantho University. Bacteria strains were isolated from diseased shrimps, pond water, and mud in the Mekong Delta, Viet Nam.
2.2. Isolation of Vibrio spp.
Bacteria were isolated from diseased shrimps, pond water, and mud in the Vinh Thuan district, Kien Giang, Bac Lieu Provinces. Diseased shrimps were collected and dissected for hepatopancreas and intestines and were homogenized in a tryptic soy broth (TSB) medium supplemented with 1.5% NaCl and spread on a thiosulfate citrate bile salt sucrose (TCBS) medium. Water and mud samples were added to a TSB medium supplemented with 1.5% NaCl, incubated for 24 h, and spread on a TCBS medium. Green colonies were selected for further study.
2.3. Identification ofV. parahaemolyticus by PCR
V. parahaemolyticus was identified using two sets of primers. The amplification of the 16S rRNA gene with 700 bp amplicons using primer F (5’-CAGGCCTAACACATGCAAGTC-3’) and primer R (5’-GCATCTGAGTGTCAGTATCTGTCC-3’) was described by Mohamed et al. (2017) for identifying Vibrio spp. Strains that possessed the needed bands were then tested for their virulence properties using the ToxR gene F (5’- GTCTTCTGACGCAATCGTTG -3’) and R (5’- ATACGAGTGGTTGCTGTCATG -3’) which is specific for V. parahaemolyticus (Kim et al., 1999). The ToxR gene was amplified with 368 bp amplicons.
2.4. Double Layer Agar Technique
The test used the Double Agar Layer Technique. Two agar layers were prepared for 2 host tests: soft agar (0.3% agar, low agar concentration) was mixed with bacteria which was incubated in 24 h and overlaid on a hard agar layer (1.5% agar). The medium used in this study was King B supplemented with 0.5% NaCl. Afterward, 1 μL of bacteriophage was inoculated and incubated for 24 h to observe the plaques. The experiment was repeated once.
2.5. Plating Method
The experiment was conducted with four treatments where the extract and bacteriophage concentrations were used for testing, and they were 8 μg/mL and 20% of 106 PFU/mL, respectively, (treatment A consisted of 100 μL of bacteria; treatment B consisted of 100 μL of bacteria and 2 μL of extract; treatment C consisted of 100 μL of bacteria and 2 μL of bacteriophages; and treatment D consisted of 100 μL of bacteria, 2 μL of extract and 2 μL of bacteriophages).
The number and the morphology of colonies were recorded after 1 h, 3 h, and 24 h. The experiment was replicated 3 times, and the Stagraphic Centurion XVIII was used to analyze the data of a number of colonies. The size of colonies was observed using the CountPHICS software.
3. RESULTS
3.1. Isolation and Identification of V. parahaemolyticus
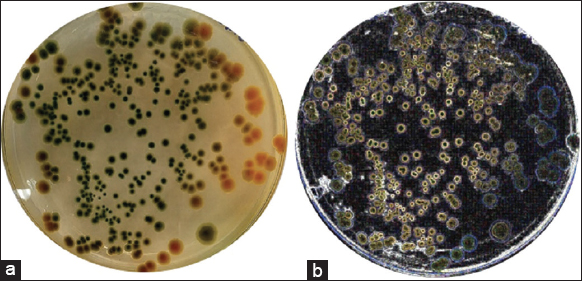
There were 48 bacterial strains that could grow on the TCBS media. All strains of bacteria were isolated from diseased shrimp with a morphological characterization similar to that of Vibrio spp. The colonies of these bacteria appeared circular, convex, and glossy. They were green in color and exhibited a diameter of about 1–2 mm on the TCBS medium after 24 h of incubation [8-10] [Figure 1]. For further confirmation, all the isolates were screened using 16S rRNA [Figure 2] and ToxR [Figure 3] primers, and were at 700 bp and 368 bp, respectively [11]. This study isolated 45 strains of Vibrio sp., including 8 strains of V. parahaemolyticus from diseased shrimp, pond water, and mud. B4XT4 and B4X0T2.2 strains were selected for the following experiments.
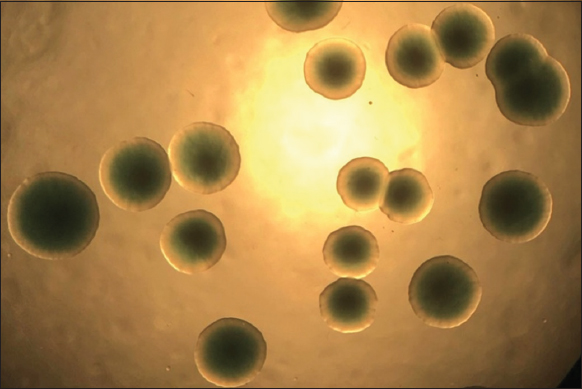 | Figure 1: Vibrio parahaemolyticus colony morphology on TCBS agar. [Click here to view] |
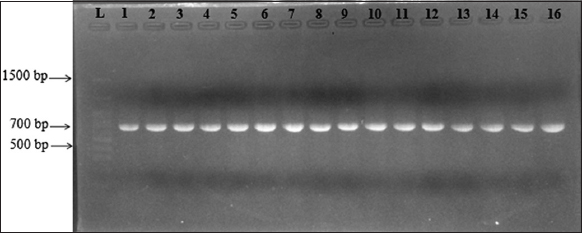 | Figure 2: Gel electrophoresis of PCR products of 16S rRNA gene. (L: 1500bp ladder; 1: B4X0T2.1; 2: B4X0T2.2; 3: B4X0T2.3; 4: V4.1; 5: B4XCT1.1; 6: B4XCT1.2; 7: V4.2; 8: B4XCT1.2; 9: B4XCT1.5; 10: V4.3; 11: B3XD2; 12: V4.6; 13: B3DX4; 14: V4.7; 15: B3XD1; 16: V4.10). [Click here to view] |
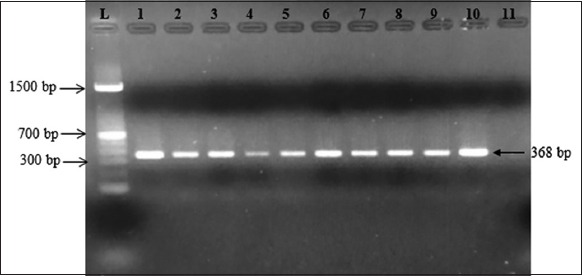 | Figure 3: Gel electrophoresis of PCR products of toxR gene (L: 1500 bp ladder; 1: positive control; 2: B4X0T2.2; 3: B4X0T2.3; 4: B3XD2; 5: V4.1; 6: V4.2; 7: V4.3; 8: V4.6; 9: V4.6; 10: V4.10; 11: negative control). [Click here to view] |
3.2. Evaluation of Host Range
The infectiveness of 5 bacteriophages against 2 strains of V. parahaemolyticus B4XT4 and V. parahaemolyticus B4X0T2.2 is noted in Table 1. It is noteworthy that phage VD3 and phage VD4 displayed the possibility of infecting both strains of V. parahaemolyticus, but strains phage VD1 and phage VD5 only infected V. parahaemolyticus B4X0T2.2. On the other hand, the strain phage VD2 could infect neither V. parahaemolyticus B4XT4 nor V. parahaemolyticus B4X0T2.2.
Table 1: Evaluation of inhibitory activity on V. parahaemolyticus of bacteriophages.
| Vibrio parahaelyticus | Phage VD1 | Phage VD2 | Phage VD3 | Phage VD4 | Phage VD5 |
|---|---|---|---|---|---|
| B4XT4* | - | - | + | + | - |
| B4X0T2.2 | + | - | + | + | + |
+: Showed the phages capable of infecting host bacteria, - sign indicates that the phages incapable of infecting host bacteria.
3.3. Number of Colony Formation
The ability of the extract and phage to inhibit the growth of V. parahaemolyticus B4XT4 and V. parahaemolyticus B4X0T2.2 was different [Figure 4] after four treatments incubated at 1 h and 3 h. The number of colonies when adding extracts or phages both increased the number of bacteria compared to certificates. For V. parahaemolyticus B4XT4, when combining the phage and extract, the inhibitory effect on bacterial growth was more obvious with only the extract (the value decreased from 7.71 to 7.21 Log CFU/mL). For V. parahaemolyticus B4X0T2.2, it was the opposite, as adding the extract activated its growth. The study which was combined phage and P. amarus extract to inhibit the growth of V. parahaemolytiucs; however, initially there was no effect.
 | Figure 4: Interaction between extract and phage on bacterial population (a) affecting Vibrio parahaemolyticus B4XT4. (b) Affecting Vibrio parahaemolyticus B4X0T2.2. [Click here to view] |
3.4. Effect of Plant Extract on the Colony of V. parahaemolyticus
Both strains of V. parahaemolyticus (B4XT4 and B4X0T2.2) were affected by the extract P. amarus, which changed the colony’s phenotype. The extract affected the shape of colonies — it caused the edge of V. parahaemolyticus B4X0T2.2 shrink and dentate with sharp and small peaks, compared to the round colonies of the control [Figure 5a and b]. The cover of the colonies was toothed, and lobate margins arose for B4XT4 [Figure 5c and d]. With regard to the colonies of V. parahaemolyticus B4X0T2.2, the size is smaller but insignificant. The most significant change in colony size was observed during a sharp decrease of 4 mm2 and 4.5 mm2, which was a drop of 1.3 times and 3.0 times, respectively [Figure 6a and b]. The decline in colony size apparently occurred in V. parahaemolyticus B4XT4; indeed, a size of 6 mm2 và 6.5 mm2 is notable, with a drop of 1.2 times and 1.1 times, respectively [Figure 6c and d].
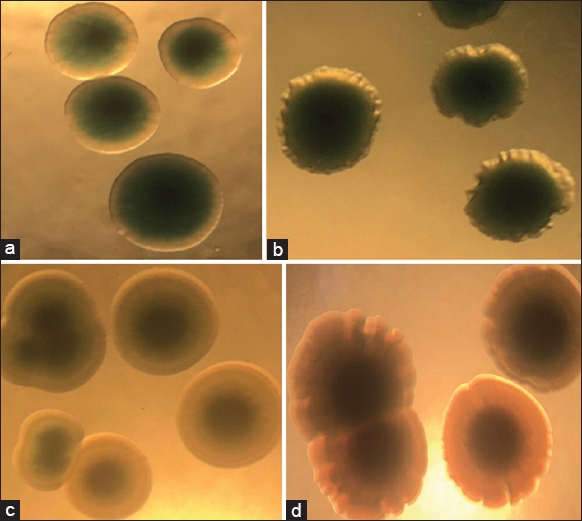 | Figure 5: Effect of plant extract on colonies of Vibrio parahaemolyticus B4XT4 and B4X0T2.2. (a) B4XT4; (b) B4XT4+ plant extract; (c) B4X0T2.2; (d) B4X0T2.2+ plant extract. [Click here to view] |
 | Figure 6: Colony size changes of Vibrio parahaemolyticus B4X0T2.2 and B4XT4 in the presence of Phyllanthus amarus extract. (a) B4X0T2.2; (b) B4X0T2.2+ plant extract; (c) B4XT4; (d) B4XT4+ plant extract. [Click here to view] |
3.5. Changes in Colonial Morphology in the Presence of Bacteriophages
The colonial morphology of V. parahaaemolyticus B4XT4 and V. parahaaemolyticus B4X0T2.2, when infected with a phage, changed compared with the control. PhageVD1 infected V. parahaemolyticus B4XT4 and V. parahaaemolyticus B4X0T2.2, causing the colony to shrink and change its serration compared to the control [Figure 7].
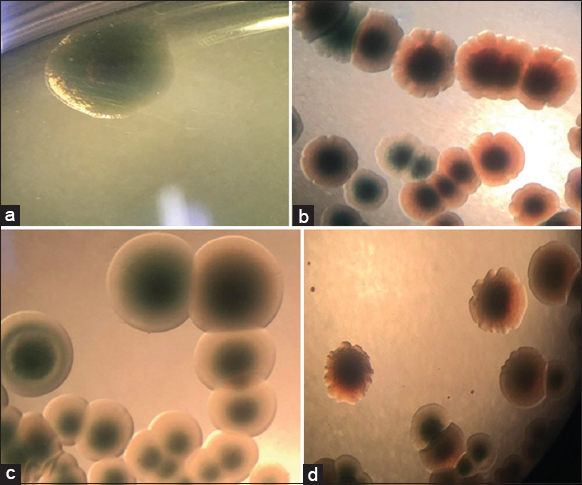 | Figure 7: VD1 changes colony phenotype and size of Vibrio parahaemolyticus B4X0T2.2 and B4XT4. (a) B4X0T2.2; (b) B4X0T2.2+phageVD3; (c) B4XT4; (d) B4XT4+phageVD1. [Click here to view] |
3.6. Impact of Plant Extract and Phages on V. parahaemolyticus
A comparison of the colony sizes in treatment between phage VD1, that could infect both strains V. parahaemolyticus, and phage VD3, which could only infect B4XT4, was conducted. As expected, the colony sizes of the former experienced a remarkable decline [Figure 8], though the latter made V. parahaemolyticus B4X0T2.2 remain the size of the colony [Figure 9].
 | Figure 8: Colony size changes Vibrio parahaemolyticus B4X0T2.2 and B4XT4 in the combination of Phyllanthus amarus extract and bacteriophage. (a) B4X0T2.2; (b) B4X0T2.2+ plant extract+phageVD1; (c) B4XT4; (d) B4XT4+ plant extract+phageVD1. [Click here to view] |
 | Figure 9: Colony size changes Vibrio parahaemolyticus B4X0T2.2 and B4XT4 in the combination of Phyllanthus amarus extract and bacteriophage. (a) B4X0T2.2; (b) B4X0T2.2+ plant extract+phageVD3; (c) B4XT4; and (d) B4XT4+ plant extract+phageVD3. [Click here to view] |
4. DISCUSSION
There have been some studies utilizing bacteriophages and P. amarus to inhibit V. parahaemolyticus but it was combined in single. Even so, all those factors presented their antimicrobacterial ability [12,13]. Unfortunately, the plant extract did not show an inhibitory effect in this experiment since the number of colonies still remained. That is possibly due to the fact that the concentration and incubation time of the extract was not optimal, so we could not conclude the efficacy of the plant extract in this regard. In addition, most of the experiments involving the use of plant extracts or bacteriophages combined with plant extracts to inhibit bacteria utilized the agar plate diffusion method to test the susceptibility of bacteria. However, this method has certain limitations — for example, molecules with a higher molecular weight diffuse very slowly in the agar, making it impossible to accurately measure the diameter of the halo ring in the plate. In contrast, in this experiment, the change of colonies was observed by using the plate count method. Although the effect of the extract on V. parahaemolyticus has not been elucidated, the colony form changed as they were affected by bacteriophages and the extracts clearly recorded the same.
Some studies on the topic of bacteriophages combined with plant extracts showed that the results were not satisfactory, except for the separate combination of the two factors [14]. Which used two types of bacteriophages (which have been identified) and three herbs combined to inhibit E. coli and concluded that it is necessary to further investigate the concentration of extracts and phages as well as combine many other ingredients to ensure the best results. The phage concentrations in this study were considered low because they were diluted to 5 times less than the original concentration. That prompted the use of the compound carvacrol, which is a compound found in many herbal essential oils (marjoram essential oil, essential oil of lemon basil, etc.), to achieve a purity rate of 99%. This only acted on but not completely inhibits the spread of the bacteria. In the experiment of [15] it was also found that the number of bacteria increased over time in the presence of bacteriophages; whereas the addition of carvacrol prevented the bacteria from becoming resistant to phages. Another critical point to note is that carvacrol is a purified substance, whereas in this experiment, we used P. amarus extract, which contains many unknown compounds that could work against the target bacteria.
Incidentally, it can be observed that there are some differences in colony morphology when treated with phages and P. amarus extract. The colony shapes were not round, but were spread out like a propeller, and such a colony form was referred to as a “competition type” [16]. That meant that the bacteria were motivated against the inhibitory factors that led to formation of the competition type. It is predicted that there existed a hidden genetic variation, which caused new phenotypes, and it was proved by True and Lindquist [17] on a yeast prion. Therefore, this experiment aimed to limit the pathogenicity of V. parahaemolyticus by making their genome not as intact as before.
In addition, a unique feature of this study is the use of the countPHICS software to measure the diameter of bacterial colonies. This software was introduced by Brzozowska et al. [18] and was supposed to be more reliable than manual measurement. The μ number in the images was higher than normal, indicating that the image resolution was low. This was due to the fact that the photos on the discs were taken only at 8 MP resolution. Fortunately, the included image processing software (ImageJ) was able to correctly evaluate the colonies for processing and for retrieving colony size data [Figure 10]. The count PHICS software was highly recommended in the colony size measurement experiments.
 | Figure 10: Images of Vibrio parahaemolyticus B4X0T2.2 before and after undertaking the ImageJ software. (a) Vibrio parahaemolyticus B4X0T2.2 before undertaking the ImageJ software. (b) Vibrio parahaemolyticus B4X0T2.2 after undertaking the ImageJ software. [Click here to view] |
5. ACKNOWLEDGMENTS
The authors would like to thank the Department of Research Affairs, Institute of Food and Biotechnology, Can Tho University for their valuable support. This study is fully funded by the Ministry of Education and Training.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
The study was funded by the Ministry of Education and Training, Vietnam (Project code: B2020-TCT-03).
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Nakai T, Sugimoto R, Park KH, Matsuoka S, Mori K, Nishioka T,
2. Tanji Y, Shimada T, Fukudomi H, Miyanaga K, Nakai Y, Unno H. Therapeutic use of phage cocktail for controlling
3. Chen L, Fan J, Yan T, Liu Q, Yuan S, Zhang H,
4. Holtappels D, Fortuna K, Lavigne R, Wagemans J. The future of phage biocontrol in integrated plant protection for sustainable crop production. Curr Opin Biotechnol 2021;68:60-71. [CrossRef]
5. Gonelimali FD, Lin J, Miao W, Xuan J, Charles F, Chen M, et al. Antimicrobial properties and mechanism of action of some plant extracts against food pathogens and spoilage microorganisms. Front Microbiol 2018;9:1639. [CrossRef]
6. Ismail CA, Deris ZZ, Bakar RA, Ismail N.
7. Paria P, Kunal SP, Behera BK, Mohapatra PK, Das A, Parida PK,
8. Hikmawati F, Susilowati A, Setyaningsih R. Colony morphology and molecular identification of
9. Tinh TH, Elayaraja S, Mabrok M, Gallantiswara PC, Vuddhakul V, Rodkhum C. Antibacterial spectrum of synthetic herbal-based polyphenols against
10. Zhang X, Sun J, Chen F, Qi H, Chen L, Sung YY,
11. Ribeiro AM, de Sousa JN, Costa LM, Oliveira FA, Santos RC,
12. Jun JW, Shin TH, Kim JH, Shin SP, Han JE, Heo GJ,
13. Phuong TV, Yen PT, Linh NQ. Antibacterial activity of extracts from dried and fresh herbal plant (
14. Pimchan T, Cooper CJ, Eumkeb G, Nilsson AS.
15. Ni P, Wang L, Deng B, Jiu S, Ma C, Zhang C,
16. Blandchard AE, Lu T. Bacterial social interactions drive the emergence of differential spatial colony structure. BMC Syst Biol 2015;9:59. [CrossRef]
17. True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 2000;407:477-83. [CrossRef]
18. Brzozowska B, Ga?ecki M, Tartas A, Ginter J, Ka?mierczak U, Lundholm L. Freeware tool for analysing numbers and sizes of cell colonies. Radiat Environ Biophys 2019;58:109-17. https://doi.org/10.1007/s00411-018-00772-z PMid:30673853 PMCid:PMC6394662