1. INTRODUCTION
Desert truffles are tuber-shaped hypogeous ascocarps of mycorrhizal Ascomycetes. They have a distinct geographical distribution restricted to semi-arid and dry portions of Saudi Arabian desert regions [1]. In the Mediterranean Basin and the Middle East, various kinds of desert truffles have been discovered [2]; however, certain uncommon species can be found in Asia [3]. The fruiting bodies of several edible hypogean fungi belonging to the phylum of Ascomycota, which were categorized in the order of Pezizales after phylogenetic studies, are generally known as desert truffles [4]. The Terfe Balsamia, Eremiomyces, Elderia, Kalahari Mycocleandia, Mattirolomyces, Phalangium, Picoa, Reddelomyces, and Ulurua genera are among those included in this category [5]. However, the only Basidiomycetes that are regarded as a desert truffle are Horakiella watarrkana [6]. The word “truffle” originates from the Latin term “tuber,” which means “swelling” in various regions of the world. “Al-Kamah” signifies “covered” in Arabic, whereas “Al-Faaga’a” pertains to the cracking of the soil above the fruiting body that occurs after it has swollen [1]. The diverse morphological keys proposed over the years are crucial [7,8]. It has been possible to establish a convincing delineation between species and taxa through molecular (gene sequencing studies) and morphological methods [9-11].
Desert truffles were discovered more than a quarter century ago and grow naturally in many parts of Saudi Arabia [12-14]. For a long time, desert truffles have been a staple food and a source of income. In addition, truffles are utilized as an edible food source because they contain a considerable number of high-quality proteins, fiber, amino acids, carbohydrates, and fatty acids [15]. Moreover, they have been used in traditional medicine for their antioxidant, anti-inflammatory, antibacterial, antifungal, antiviral, and anticancer properties [3,16]. Recently, they have been used for treating eye pathogens [17]. Saudi Arabians have a long history of collecting desert truffles in natural conditions. As a result of the ongoing overexploitation of this valuable natural resource, this crop is at risk, underscoring the need for its protection. Two types of Saudi Arabian truffles are also present. They can be distinguished by their harvesting area, size, color, and host. “Terfess red of Tafilalet,” “Terfess white of Tafilalet,” “Ezzebdi,” or “Terfess rose of Mamora” are examples of these terms. Each type appears in the rainfall season in spring, although some can emerge as early as November and December. Hafr Al Batin region’s desert truffle has received very little research, so determining its diversity must be considered in the context of Saudi Arabia’s fungal diversity.
The majority of Terfezia species form mycorrhizal symbioses with Cistaceae plants, primarily with perennial and annual Helianthemum species [18-20]. This mycorrhizal association is extremely well adapted to semi-arid environments by exploiting a physiological mechanism to resist drought [21-23]. The significant variation in the soils found in desert truffles reflects the environmental settings in which they grow. There is evidence that Terfezia species can adapt to a wide range of soil pH, edaphic conditions, and textures [24]. The discovery of new species is made possible using cutting-edge molecular techniques on the hypogeous fungus group, which is home to desert truffles. The number of new species discovered within the genera of desert truffles will rise [25]. Two Terfezia isolates were the primary focus of this study’s description. To achieve this, we conducted conventional morphological studies along with phylogenetic analysis using Internal Transcribed Spacer (ITS)-rDNA sequences from Terfezia samples collected from Hafr Al Batin, Saudi Arabia.
2. MATERIALs AND METHODS
2.1. Study Area
Hafr Al Batin (28°26′3″N 45°57′49″E), located in the eastern region of Saudi Arabia, has an arid climate, and the annual rainfall is 63 mm [26]. The average temperature from December to February ranges from 20–23°C as a maximum) to 6–8°C (as a minimum) and the soil type is arid soil, sand, and gravel [27].
2.2. Choice of Sampling Sites
During February to May 2022, a total of 60 fresh fruiting truffle bodies that were in full bloom were harvested. They were gathered in accordance with the recommendations and guidelines provided by individuals responsible for information gathering in the study regions. It is most common to find the Saudi Arabian truffle in the vicinity of Helianthemum herbaceous plants, with mycorrhizal associations are formed by them [20,28]. Truffles were discovered in situ near the roots in the species Helianthemum [29] by examining fractures and swellings in soil surfaces induced by ascocarp development.
2.3. Samples Preparation
Samples were divided into two groups (white and brown). Eight gleba cubes of each group were sampled and stored in a 1.5 mL Eppendorf tube containing 70% ethanol (v/v) to preserve the DNA for further analysis. At the same time, a portion of fresh gleba was scraped with a scalpel from each truffle and mounted on a microscopic slide with glycerol to perform spore morphological analysis. The morphological characterization of truffle spores was conducted using a light microscope (Euromex iScope), and photographs were taken with an OLYMPUS DP74 digital camera. Genetic analysis was employed to compare molecular information with the other available data.
2.4. Morphological and Microscopic Characteristics
The morphology of macroscopic features, particularly shape and color, was detailed for each portion of the truffle, namely the ascocarp and the gleba. Samples were sliced using a sharp blade, immersed in 5% KOH, and examined with an optical microscope (×100). Various properties such as ascospore form, quantity per ascus, and position were assessed for species identification [7,30,31]. All this information was compared to that previously described by various authors [9,10,32-35].
2.5. Analysis of Soil Samples
Soil samples were collected at a depth of approximately 10–25 cm at each truffle harvesting location. At a temperature of 25°C in the open space, soil samples were air-dried in the laboratory. To evaluate the samples’ physical and chemical properties, they were sieved through a 2 mm sieve and then subjected to standard analysis techniques. As part of the analysis, soil parameters such as particle diameter, electrical properties, pH, and total CaCO3 were assessed. Salinization was determined using the following equations: 640 conductivities in dS/m [36]; pH was measured with a pH-meter on a soil: water ratio of 1:5; and total CaCO3 was assessed by Bernard calcimeter. Phosphorus, Ca++, Mg++, and K+ were measured by an atomic absorption spectrometer on a soil: water extract ratio of 1:5 [36]. HCO3− was titrated with H2SO4 and SO4 by the gravimetric analysis following precipitation as barium chloride. Using sterile water and 1 g of dry weight, the soil sample was processed into a suspension for microbiological examination. To estimate aerobic heterotrophic fungal populations, the usual spread-plate dilution method published by Seeley and VanDemark [37] was employed. One mL of the soil suspension was diluted serially (10 times). To isolate the fungus, potato dextrose agar was supplemented with 0.05% (w/v) chloramphenicol (to prevent bacterial growth) and incubated at room temperature for 7 days.
2.6. Molecular Characterization
2.6.1. DNA extraction
Genomic DNA was extracted from the two isolates following the methods of Hassan et al. 2022 [38]. The samples were ground with liquid nitrogen and homogenized in a sterilized mortar with 1.5 mL of CTAB extraction buffer, 20 μL of Triton x-100, and 100 μL of 10% SDS. The sample mixture was transferred to a microcentrifuge tube and incubated at 60°C for 60 min. The supernatant from the samples was transferred to a new Eppendorf tube after being centrifuged at 15,000 rpm for 10 min. The supernatant was treated with an equal volume of chloroform-isoamyl alcohol before being centrifuged at 10,000 rpm for 10 min. The aqueous phase was transferred gently to a new Eppendorf tube. To precipitate in nucleic acids, 0.45 volumes of ice isopropanol were added and mixed thoroughly by inversion before being incubated at 25°C for 1 h. Following incubation, samples underwent a 15-min, 15,000 rpm centrifugation. After discarding the supernatant, the pellet was washed with 1 mL of 70% ethanol and centrifuged at 12,000 g for 5 min. The pellet underwent a 5-min, room-temperature air drying process. Then, the elution step was performed using 50 μL of TE buffer. The concentration and purity of purified DNA were measured by BioTek Epoch2 Microplate reader (Thermo Scientific, Waltham, MA, USA). DNA purity was >1.8 ± 0.20 under the absorbance ratio A260/A280.
2.6.2. Polymerase chain reaction (PCR) assay
Based on fungal genomic DNA, PCR amplification was carried out utilizing the ITS region of the rDNA, which contains the 5.8S ribosomal gene. The universal primer pairs ITS 1 (TCCGTAGGTGAACCTTGCGG) and ITS 4 (TCCTCCGCTTATTGATATGC) were used for PCR amplification [39]. PCR reaction was performed in a 25 μL mixture containing 0.2 μM of each primer at a concentration of 10 pM, 200 μM of dNTPs mix, 2.5 μL of ×10 PCR reaction buffer, 1.5 μM MgCl2, 1.25 units of TAKARA Taq DNA polymerase (Cat. #: R001AM), 2 μL of template DNA, and the final volume was adjusted with sterilized double-distilled water. The reactions were amplified using a PCR thermocycler (AriaMx), which had the following settings: 95°C for 3 min, 35 cycles at 95°C for 50 s, 52°C as the annealing temperature for 1 min, an extension of 72°C for 1 min, and a final extension temperature of 72°C for 10 min. For further purification and downstream use, amplified PCR products were stored at 20°C.
2.6.3. DNA sequencing
PCR amplicons were purified according to the manufacturer’s QIAquick Gel Extraction Kit (Cat. #: 28704). The purified PCR fragments were sequenced by Macrogen Company, South Korea, using the automated Sanger Sequencing method. The obtained sequences for the ITS-rDNA gene were analyzed using the Standard Nucleotide BLAST tool and registered in the NCBI database under accession numbers ON909007 and ON909008 for Terfezia claveryi isolate Hafr Al Batin and Hafr Al Batin2, respectively. (http://www.ncbi.nlm.nih.gov).
2.6.4. Pairwise alignment and phylogenetic construction
Jalview software [40] was utilized to display single-nucleotide polymorphisms (SNPs) and the consensus generated from the pairwise sequence alignment of the two obtained isolates, as well as their alignment with the nearest strains in the NCBI database. (http://www.jalview.org/).
DNASTAR (version 5.05) was employed in this analysis to generate contiguous sequences of the Terfezia isolates. Sequences from the most closely related species within the Terfezia genus were retrieved from GenBank. The Peziza montirivicola strain M 0274465 (NR_148194; type material) was used as the outgroup for the phylogenetic tree in this study. MAFFT (Katoh and Standley, 2013) [41] was used to align all sequences, while BMGE (Criscuolo and Gribaldo, 2010) [42] was used to refine the alignment by addressing gaps and removing weak, uninformative characters. Phylogenetic analyses utilizing maximum-likelihood (ML) and maximum-parsimony (MP) methodologies were performed using MEGA X (version 10.2.6) (Kumar et al., 2018) [43]. The robustness of the most parsimonious trees was evaluated through 1,000 replications (Felsenstein, 1985) [44]. The optimal nucleotide substitution model for the ML analysis was determined using the Akaike Information Criterion in ModelTest 3.7 (Posada and Crandall, 1998) [45].
3. RESULTS AND DISCUSSION
3.1. Truffle Presence and Distribution
Desert truffles are only available in a few locations, such as Turkey, Portugal, Spain, France, and other nations in the Mediterranean basin [46-48]. Terfezia and Tirmania are the two genera of the harvested Terfez. Four species make up the genus Terfezia: Terfezia olbiensis, T. leptoderma, T. boudieri, and T. claveryi. One species, Tirmania nivea, belongs to the genus Tirmania. From February to May 2022, the author sampled various locations in the Hafr Al Batin region and collected a total of 60 fresh fruiting truffle bodies. The harvested truffle ascocarps typically have a spherical appearance, a lobed shape, and a foot (pedicel) at the base. The surface of the peridium is quite smooth and covered with tiny scales, as observed under a binocular magnification for collected types. Small pale yellowish veins, cream color, and white contribute to the distinctive appearance of the gleba.
The first isolate, T. claveryi 49–621, was the most widely distributed species across all surveyed sites, with a mean relative abundance of 63.4%, followed by the second isolate, T. claveryi 1–564, whose incidence varied at 11.7%. Harvested species were associated with H. lipii in the Hafr Al Batin region. Our results indicated that the desert truffles grow in the sand, near Helianthemum roots. This result agrees with Bermaki et al., 2017 [28].
3.2. Study of the Morphology of the Examined Truffles
T. claveryi 1–564 - harvested February 2021. Up to 11.6 × 4.7 cm in size, ascocarps that are frequently subglobose in shape, occasionally flattened, and possess a short, smooth, or wrinkled pedicel that is white to cream in color before turning yellow [Table 1A]. The peridium is between 0.5 and 2 mm thick and has a light cream tint that turns white when exposed to sunlight. The gleba is whitish or yellow, squishy, and dotted with tiny veins. When examined under a microscope, T. claveryi’s gleba reveals numerous hyphae containing ascospores. Asci [Table 1C] are pedicelled, ellipsoid to ovoid in shape, and range in size from 62 to 78 × 33 to 42 μm. They typically feature 6 amyloid ascospores. Their pedicle is short and has a thin wall.
Table 1: Some morphological and anatomical features of fruiting bodies of Terfezia claveryi 1–564 (A: Ascocarp, C: Ascus with six ascospores) and Terfezia claveryi 49–621 (B: Ascocarp, D: Ascus with eight ascospores), collected from the Hafr Al Batin region.
| Sample ID/Query Accession Number | Recognized Species | Collection Date/site/Host plant | Ascocarps | Ascus and Spore Morphology | Consensus Taxa/GenBank Accession Number |
|---|---|---|---|---|---|
| 1–564/ON909007 | Terfezia claveryi | February 2022/Hafr Al Batin/H. lippii |  | 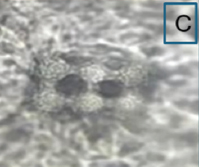 | MK967454.1/Iraq |
| 49–621/ON909008 | Terfezia claveryi | April 2022/Hafr Al Batin/H. lippii |  |  | GQ888692.1/Iran |
T. claveryi 49–621 was harvested in April 2021. The peduncle at the base of subglobose to tuber-form ascocarps measures 5.9 × 12.4 cm in diameter [Table 1B]. Its surface ranges from dark brown to brown and is cracked. Peridium: Brownish, 0.75–2 mm thick, with a smooth, often fractured surface. The gelatinous skin has obvious veins that irregularly define fertile patches, reddish to pinkish in color. Asci: 62–78 × 71–79 μm in diameter, subglobose or ovoid, randomly distributed in fungal tissues. It contains eight ascospores when fully mature. The spherical yellowish-hyaline ascospores measure 19–22 × 21–24 μm, floating freely inside asci, with thick double-walled structures and a germ pore [Table 1D]. The morphological and anatomical characteristics of T. claveryi relevant to this study perfectly align with previous characterizations of the same truffle species (Moracco [49]; Tunis [2]; Kuwait [50]; Saudi Arabia [1]; Iraq [51]; and Egypt [52]).
3.3. Soil Analysis
Table 2 presents the results of the microbiological and physicochemical studies conducted on soil samples collected from various locations in the Hafr Al Batin region. Soil investigations of desert truffle-producing locations revealed significant homogeneity in pedological properties across the two truffle isolates. A study of the soils indicated a sand-like texture, extremely low amounts of organic matter, an alkaline pH, and very low levels of nitrogen and phosphate. Conversely, the amounts of potassium and calcium were high. According to a study on grain size based on the American system of classifying soil textures, all the habitats for truffles in the Hafr Al Batin have soils with a sandy texture and single-grained structure, exhibiting high rates of sand grains (72.7–73.2%). This sample had a sand-like texture, minimal organic matter, an alkaline pH, and exceptionally low nitrogen and phosphorus contents. Low levels of organic matter are closely associated with low soil P availability [24]. It was found that the pH range of desert truffle habitats was between 8.1 and 8.3. According to the 1:5 aqueous extract’s scale of categorization for salinity, the electrical conductivity values were low and ranged between 0.82 and 0.85 dS/m, indicating non-saline soils. Organic materials constituted up 0.71–0.83% of the total substance. These results demonstrate a significant deficiency in soil organic matter, as measured by Morond’s scale of organic matter [53,54]. Researchers have found that desert truffles form symbiotic relationships with Cistaceae roots, particularly Helianthemum lippii. The genus Helianthemum is well-known in literature for establishing connections with truffles in many parts of the world, such as Algeria [33] and Moracco [28].
Table 2: Microbiological and physiochemical soil analyses at desert truffle harvesting sites in the Hafr Al Batin region.
| Soil Parameter | Terfezia claveryi 49–621 (n=16) | Terfezia claveryi 1–564 (n=7) |
|---|---|---|
| Chemical composition | ||
| Organic matter (%) | 0.83 | 0.71 |
| K+ (cmol+/kg) | 0.16 | 0.16 |
| Phosphorus (ppm) | 19.18 | 19.95 |
| Ca++ (cmol+/kg) | 8.11 | 8.38 |
| Mg++ (cmol+/kg) | 2.05 | 1.99 |
| HCO3− (cmol+/kg) | 0.46 | 0.32 |
| SO4− (cmol+/kg) | 0.32 | 0.33 |
| Texture (%) | ||
| Clay | 1.9 | 1.7 |
| Silt | 9.5 | 9.2 |
| Sand | 73.2 | 72.7 |
| Ec (dS/m) | 0.85 | 0.82 |
| pH | 8.3 | 8.1 |
| Microbiological analysis (cfu/gm soil) | ||
| Mucor circinelloides van Tiegham | 8.6×105 | 7.3×105 |
3.4. DNA Sequences and Phylogenetic Analyses
The amplified PCR products of the ITS-rDNA gene of the isolates were sequenced and deposited in the NCBI database under accession numbers ON909007 and ON909008 for T. claveryi isolate Hafr Al Batin and Hafr Al Batin 2, respectively. BlastN and Jalview alignment results revealed that the two isolates, Hafr Al Batin and Hafr Al Batin 2 were closely like T. claveryi strain MZR12 (MK967454) and T. claveryi isolate Kh1 (GQ888692) with identity ratios of 97.34% and 98.74%, respectively. Pairwise alignment analysis of T. claveryi isolate Hafr Al Batin (ON909007) and the nearest sequence (MK967454) exhibited 8 SNPs and 2 GAPs [Figure 1]. Meanwhile, the alignment between T. claveryi isolates Hafr Al Batin2 (ON909008) and the nearest sequence (GQ888692) revealed 5 SNPs and one gap [Figure 2]. Interestingly, the pairwise sequence alignment between the two obtained isolates indicated a 97.1% identity ratio. This divergence resulted from 9 SNPs and 3 gaps, reflecting possible different features among the two isolates [Figure 3].
 | Figure 1: The image shows single-nucleotide polymorphisms and GAPs between Terfezia claveryi isolate Hafr Al Batin (ON909007) and the nearest sequence (MK967454) deposited in GenBank. [Click here to view] |
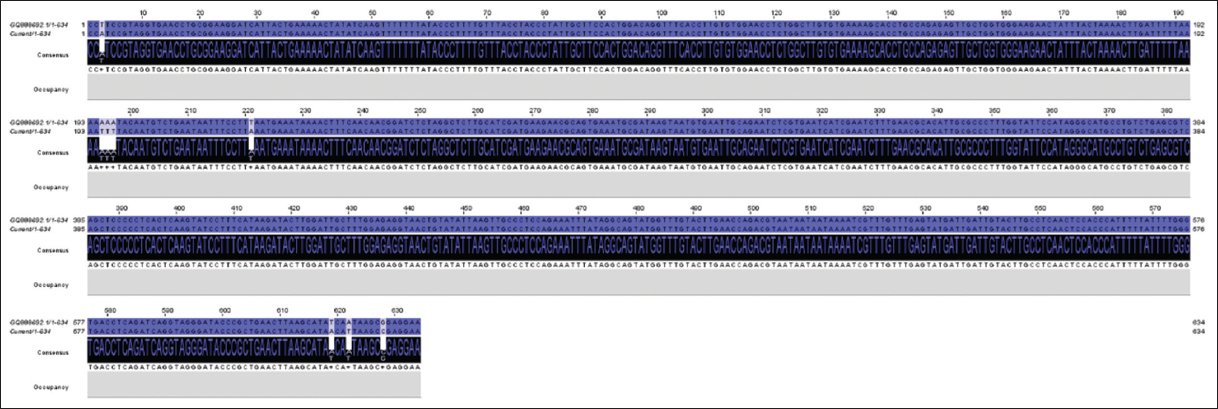 | Figure 2: The image shows single-nucleotide polymorphisms and GAPs between Terfezia claveryi isolate Hafr Al Batin2 (ON909008) and the nearest sequence (GQ888692) deposited in GenBank. [Click here to view] |
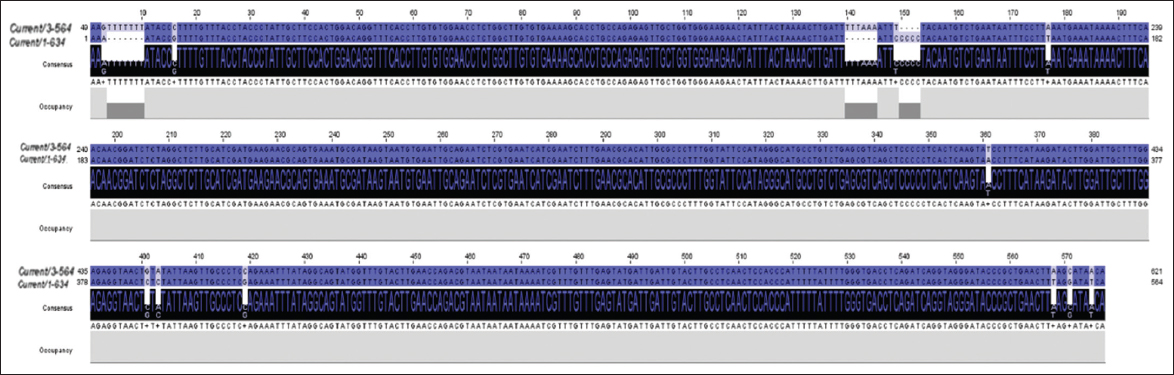 | Figure 3: The image shows single-nucleotide polymorphisms and GAPs between the two obtained isolates Hafr Al Batin (ON909007) and Hafr Al Batin2 (ON909008). [Click here to view] |
In phylogenetic analysis, thirteen sequences produced a total of 614 characters in the final ITS dataset, of which 442 were accurately aligned, 144 were designated as variable, and 57 were identified as relevant. Tamura’s 3-parameter model, utilizing a discrete gamma distribution (T92 + G), effectively illustrated the relationships among taxa. Eight trees were generated using the MP method. The ML tree [Figure 4] exhibits the following attributes: a tree length of 337, a maximum log likelihood of −2359.15, a consistency index of 0.748634, a retention value of 0.799127, and a composite index of 0.598253. The strains in this study are positioned within the same clade as T. claveryi strains RUF-TC2, J0297, MA: FU 8240, and MUB: Fung-0922, which is supported by a robust bootstrap value of 81% ML/87% MP. As a result, the isolates in this study were identified as T. claveryi [Figure 4]. The ITS sequences of T. claveryi strains in this investigation were submitted to GenBank as ON909007 and ON909008.
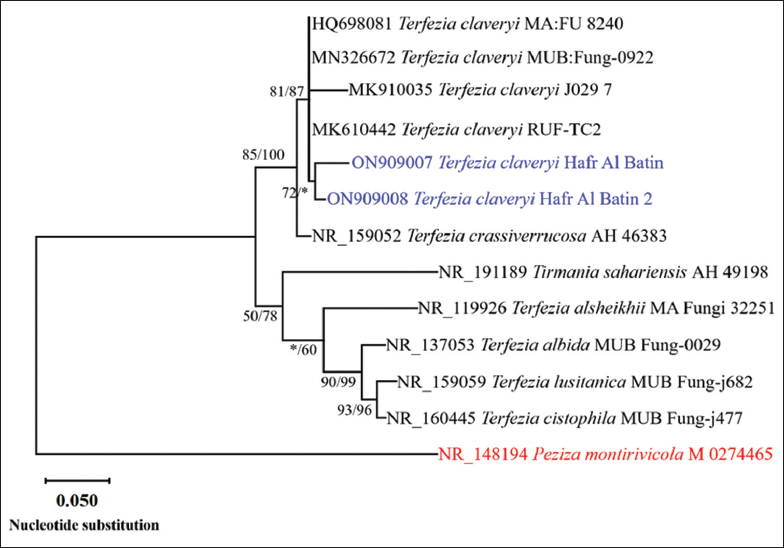 | Figure 4: The maximum likelihood evolutionary tree obtained from a heuristic search (1000 replications) using maximum-likelihood (ML)/maximum-parsimony (MP) analysis of Internal Transcribed Spacer sequences of Terfezia claveryi strains in this study (in blue) compared to the most similar species of the genus Terfezia in GenBank. Bootstrap support values for ML/MP ≥50% are indicated near the respective nodes. The tree is rooted to Peziza montirivicola M 0274465 (in red). [Click here to view] |
The morphological features of T. claveryi [Table 1] and the molecular study results [Figure 4] provide compelling evidence that these two species are one and the same. We identified these isolates as belonging to the same species by comparing them to all other taxa that were morphologically and phylogenetically comparable to them. Host specialization and edaphic tolerances (fungal and/or host tolerances) may also be important contributors to the Terfezia genus’s high species diversity, although other variables may also be at play [55]. In this regard, T. claveryi and T. albida are similar in that they both prefer alkaline soils and Helianthemum species as hosts, although the latter’s gleba is less grayish in contrast to that of T. claveryi.
4. CONCLUSION
In this taxonomic and bioecological investigation of desert truffles, which was originally conducted in Hafr Al Batin region, 2 isolates of T. claveryi have been identified. These isolates are somewhat limited, but they exhibit remarkable adaptations to their environment, characterized by severe aridity and rudimentary soil properties, which are associated with H. lippii (Cistaceae). Harvested truffles grow ecologically in sandy, slightly alkaline, somewhat calcareous soils that are low in organic and mineral content. As far as climatic conditions are concerned, these fungi occur in hot arid environments, with heavy rains, followed thereafter by periods of dryness. The ascospore sizes produced by the two isolates studied here vary. It is believed that they represent separate species since the first isolate generates spiny ascospores while the other hyaline ascospores. Our rigorous comparisons of ascocarps and ascospores, along with thorough ITS-rDNA sequence analysis, support the identification of just one species.
5. ACKNOWLEDGMENTS
The author would like to thank Professor Dr. Ahmed Moharam, Moubasher Mycological Center, Assiut University (AUMMC), Egypt, for his assistance with fungal identification.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The author reports no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Bokhary HA. Desert truffle Al-Kamah of the Kingdom of Saudi Arabia. I. Occurrence, identification and distribution. Arab Gulf J Sci Res 1987;987:245-55.
2. Slama A, Fortas Z, Neffati M, Khabar L, Boudabous A. Taxonomic study of some hypogeous Acsomycota (Terfeziaceae) from southern Tunisia. Bull Soc Mycol Fr 2006;122:187-95.
3. Owaid MN. Bioecology and uses of desert truffles (Pezizales) in the Middle East. Walailak J Sci Technol 2017;15:179-88. [CrossRef]
4. O'Donnell K, Cigelnik E, Weber NS, Trappe JM. Phylogenetic relationships among ascomycetous truffles and the true and false morels inferred from 18s and 28s ribosomal DNA sequence analysis. Mycologia 1997;89:48. [CrossRef]
5. Morte A, Honrubia M, Gutierrez A. Biotechnology and cultivation of desert truffles. In:Varma A, editor. Mycorrhiza:State of the Art Genetics and Molecular Biology, Eco-function, Biotechnology, Eco-Physiology, Structure and Systematics. Berlin, Heidelberg:Springer Verlag;2008. 467-83.
6. Trappe JM, Kovács GM, Claridge AW. Comparative taxonomy of desert truffles of the Australian outback and the African Kalahari. Mycol Prog 2009;9:131-43. [CrossRef]
7. Trappe JM. The orders, families, and genera of hypogeous Ascomycotina (truffles and their relatives). Mycotaxon 1979;9:297-340.
8. Castellano MA, Trappe JM, Maser Z, Maser C. Key to Spores of the Genera of Hypogenous Fungi of North Temperate Forests. Eureka:Mad River Press;1989. 184.
9. Iotti M, Amicucci A, Stocchi V, Zambonelli A. Morphological and molecular characterization of mycelia of some Tuberspecies in pure culture. New Phytol 2002;155:499-505. [CrossRef]
10. Marozzi G, Benucci GM, Suriano E, Sitta N, Raggi L, Lancioni H, et al. Tuber mesentericum and Tuber aestivum Truffles:New insights based on morphological and phylogenetic analyses. Diversity 2020;12:349. [CrossRef]
11. Aish AA, Saad T, Abdulmalek TA, Kareem LB, Yasir OM. Molecular identification and genetic diversity study of the Iraqi truffles. J Phytol 2020;12:121-6. [CrossRef]
12. Al-Qarawi AA, Mridha MA. Status and need of research on desert truffles in Saudi Arabia. J Pure Appl Microbiol 2012;6:1051-62.
13. Bokhary HA, Parvez S. Occurrence of Desert Truffle in Harrat Al-Harra (Northern Region), Saudi Arabia. A Survey Report Submitted to National Commission for Wildlife Conservation and Development (NCWCD), Riyadh;1987.
14. Bokhary HA, Parvez S. Desert truffle “Al-Kamah”of the kingdom of Saudi Arabia. Additional contribution. Arab Gulf J Sci Res Agric Biol Sci1988;6:103-12.
15. El Enshasy H, Elsayed EA, Aziz R, Wadaan MA. Mushrooms and truffles:Historical biofactories for complementary medicine in Africa and in the Middle East. Evid Based Complement Alternat Med 2013;2013:620451. [CrossRef]
16. Veeraraghavan VP, Sardar H, Janardhana PB, Lavina D, Malathi K, Jenifer MA, et al. A comprehensive and critical review on ethnopharmacological importance of desert truffles:Terfezia claveryi, Terfezia boudieri, and Tirmania nivea. Food Rev Int 2022;38:Suppl 1:846-65. [CrossRef]
17. Mostafa SM. Saudi practice of consuming Terfezia claveryi truffles intended for treating eye pathogens. J Clin Case Rep Stud 2024;5(9);DOI:10.31579/2690-8808/223. [CrossRef]
18. Gutiérrez A, Morte A, Honrubia M. Morphological characterization of the mycorrhiza formed by Helianthemum almeriense Pau with Terfezia claveryi Chatin and Picoa lefebvrei (Pat.) Maire. Mycorrhiza 2003;13:299-307. [CrossRef]
19. Morte A, Andrino A. Domestication:Preparation of mycorrhizal seedlings. In:Kagan-Zur V, Roth-Bejerano N, Sitrit Y, Morte A, editors. Desert Truffles. Berlin, Heidelberg:Springer;2014. 343-65. [CrossRef]
20. Alshammari S, Mahmoud S. Bioactive compounds of methanolic extract of Helianthemum lippii grows in Hafr Al-Batin region, northeastern Saudi Arabia. Acta fytotechn. Zootechny 2022;25:60-6. [CrossRef]
21. Morte A, Lovisolo C, Schubert A. Effect of drought stress on growth and water relations of the mycorrhizal association Helianthemum almeriense-Terfezia claveryi. Mycorrhiza 2000;10:115-9. [CrossRef]
22. Morte A, Navarro-Ródenas A, Nicolás E. Physiological parameters of desert truffle mycorrhizal Helianthemun almeriense plants cultivated in orchards under water deficit conditions. Symbiosis 2010;52:133-9. [CrossRef]
23. Turgeman T, Ben Asher J, Roth-Bejerano N, Kagan-Zur V, Kapulnik Y, Sitrit Y. Mycorrhizal association between the desert truffle Terfezia boudieri and Helianthemum sessiliflorum alters plant physiology and fitness to arid conditions. Mycorrhiza 2011;21:623-30. [CrossRef]
24. Bonifacio E, Morte A. Soil properties. In:Kagan-Zur V, Roth-Bejerano N, Sitrit Y, Morte A, editors. Desert Truffles. Berlin, Heidelberg:Springer;2014. 57-67. [CrossRef]
25. Bordallo JJ, Rodriguez A. Cryptic and new species. In:Kagan-Zur V, Roth-Bejerano N, Sitrit Y, Morte A, editors. Desert Truffles. Berlin, Heidelberg:Springer;2014. 39-53.
26. Sharif H, Al-Zahrani M, Hassan A. Physically, fully-distributed hydrologic simulations driven by GPM satellite rainfall over an urbanizing arid catchment in Saudi Arabia. Water 2017;9:163. [CrossRef]
27. Khan MA, Khan ST. Microbial communities and their predictive functional profiles in the arid soil of Saudi Arabia. Soil 2020;6:513-21. [CrossRef]
28. Bermaki FZ, Khabar L, Ezzanega A. Bioecology of desert truffles in the province of Figuig in Eastern Morocco. Sci Fungorum 2017;46:29-36. [CrossRef]
29. Alsheikh AM. Taxonomy and Mycorrhizal Ecology of the Desert Truffles in the Genus Terfezia. Ph.D. Thesis, Oregon State University, Oregon, USA;1994. Available from:https://file:///c:/users/dr.sabary/downloads/alsheikhabdulmagidm1995.pdf
30. Ferdman Y, Aviram S, Roth-Bejerano N, Trappe JM, Kagan-Zur V. Phylogenetic studies of Terfezia pfeilii and Choiromyces echinulatus (Pezizales) support new genera for southern African truffles:Kalaharituber and Eremiomyces. Mycol Res 2005;109:237-45. [CrossRef]
31. Khabar L. Mediterranean basin:North Africa. In:Kagan VZ, editor. Desert Truffles, Soil Biology. Berlin:Spring-Verlag;2014. 143-58. [CrossRef]
32. Segneanu AE, Sfirloaga P, David I, Balcu I, Grozescu I. Characterization of truffles using electrochemical and analytical methods. Digest J Nanomater Biostructures 2012;7:199-205.
33. Bradai L, Bissati S, Chenchouni H. Desert truffles of the North Algerian Sahara:Diversity and bioecology. Emir J Food Agric 2014;26:425-35. [CrossRef]
34. El Akil M, Ali O, Amina OT, Rachid B, Allal D. Study of eastern Morocco desert truffles. Int J Curr Res 2016;8:33922-9.
35. Kumar LM, Smith ME, Nouhra ER, Orihara T, Sandoval Leiva P, Pfister DH, et al. A molecular and morphological re-examination of the generic limits of truffles in the tarzetta-geopyxis lineage-Densocarpa, Hydnocystis, and Paurocotylis. Fungal Biol 2017;121:264-84. [CrossRef]
36. Baize D. Guide Des Analyses En Pédologie:Choix, Expression, Présentation, Interprétation. Ed. Quae. France;2000.
37. Seeley HW, VanDemark PJ. Microbes in Action. A Laboratory Manual of Microbiology. 3rd ed. USA:W.H Freeman and Company;1981. 350.
38. Hassan EO, Shoala T, Attia AM, Badr OA, Mahmoud SY, Farrag ES, et al. Chitosan and nano-chitosan for management of Harpophora maydis:Approaches for investigating antifungal activity, pathogenicity, maize-resistant lines, and molecular diagnosis of plant infection. J Fungi 2022;8:509. [CrossRef]
39. White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In:PCR Protocols. United States:Academic Press;1990. 315-22. [CrossRef]
40. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009;25:1189-91. [CrossRef]
41. Katoh K, Standley DM. Mafft multiple sequence alignment software version 7:Improvements in performance and usability. Mol Biol Evol 2013;30:772-80. [CrossRef]
42. Criscuolo A, Gribaldo S. Bmge (block mapping and gathering with entropy):A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol 2010;10:210. [CrossRef]
43. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547-9. [CrossRef]
44. Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution 1985;39:783-91. [CrossRef]
45. Posada D, Crandall KA. Modeltest:Testing the model of DNA substitution. Bioinformatics 1998;14:817-8. [CrossRef]
46. Honrubia M, Cano A, Molina-NiñC. Hypogenous fungi from southern Spanish semi-arid lands. Persoonia 1992;14:647-53.
47. Moreno G, Díez J, Manjón JL. Picoa lefebvrei and Tirmania nivea, two rare hypogeous fungi from Spain. Mycol Res 2000;104:378-81. [CrossRef]
48. Kaddouri H, Ouahmane L, Tounsi A. Desert truffle biodiversity, biology, ecology, and mycorrhizal connection in Morocco. Notulae Sci Biol 2024;16:11565. [CrossRef]
49. Khabar L, Najim L, Janex-Favre MC, Parguey-Leduc A. Contribution to the study of the mycological flora of Morocco:Moroccan truffles (Discomycetes). Extrait du Bull. Soc Mycol Fr 2001;117:213-29.
50. Alsheikh AM, Trappe JM. Desert truffles:The genus Tirmania. Trans Br Mycol Soc 1983;81:83-90. [CrossRef]
51. Owaid MN. Biodiversity and bioecology of Iraqi desert truffles (Pezizaceae) during season 2014. J Arid Agric 2016;2:22-5. [CrossRef]
52. Sallam FE, El-mokadem MT, Mekawey AA, Saker EA. Egyptian truffles as a source of antimicrobial and antioxidant agents. J Sci Res Sci 2022;39:49-61. [CrossRef]
53. Morond DT. Soil Landscape of the Woodburn 1:100,000 Sheet. Sydney:Department of Land and Water Conservation;2001. 271-6.
54. Hakkou S, Sabir M, Machouri N. Contribution to the study of the production factors of Moroccan truffles (Soil, Climate, Vegetation). J Mater Environ Sci 2023;14:13-30.
55. Díez J, Manjón, JL, Martin F. Molecular phylogeny of the mycorrhizal desert truffles (Terfezia and Tirmania), host specificity and edaphic tolerance. Mycologia 2002;94:247-9. [CrossRef]