1. INTRODUCTION
Enzymes are the essential bio-molecules that have intrigued researchers due to their outstanding properties such as catalysis and sustainability. Enzymes are used in many eco-friendly industrial applications, as they are selective and efficient. In addition, the biochemical reaction is accelerated by the use of enzymes as they lower the activation energy of the reaction [1]. Proteases are one of the major industrial enzymes and find various applications in the laundry industry, leather industry, pharmaceutical industry, food industry, cosmetics, silk degumming, silver recovery, chemical industry, and waste management processes [2,3]. Proteases can be produced from several sources such as plants, animals, and microorganisms (bacteria, yeasts, fungi, and actinomycetes). Among them, the microbial sources are renewable. The researchers have mainly studied the Bacillus species for the production of protease enzyme [4]. Microbial sources are preferred due to their fast growth in a confined space and ease in genetic manipulation to create novel enzymes with enhanced properties for various applications [5]. Studies have shown that protein engineering has been used to boost up the stability attribute of protease enzymes to develop cost-efficient strategies for large-scale applications of the enzyme [3]. The protease production from bacteria depends on the type of strain, the composition of the medium, method of cultivation, nutrient requirements, cell growth, time of incubation, pH, metal ions, temperature, and thermostability. The enzymes derived from Bacillus species are highly active and stable at different ranges of temperature and pH; purification can be easily done at a low cost [6]. Various Bacillus strains such as Bacillus pumilus [7], Bacillus horikoshii [8], Bacillus clausii [9], Bacillus subtilis [10], Bacillus sphaericus [11], Bacillus aquimaris [12], Bacillus pseudofirmus [13], and Bacillus circulans [14] are known to produce industrially important proteases. Properties such as thermostability and pH stability are important for industrial enzymes as they aid in reducing microbial contaminants as well as operational costs. Proteases stable at high temperatures and pH are used to study the stability in extreme conditions and understand the mechanism [15]. Therefore, the search for a protease with desirable properties for commercial viability is imperative [16].
Optimization of microbial fermentation is critical for improved cost-effective protease production as fermentation parameters play an important role in supporting the growth of microorganisms for enzyme production. A classical method, that is, one factor at a time (OFAT) is frequently used in the optimization process to achieve optimum cell density and high productivity. The method becomes laborious as it requires more time and experimental data sets. Besides, the interaction between the variables cannot be determined using OFAT [2]. Researchers have designed statistical experimental tools where interaction between multiple variables can be optimized by screening the suitable media components to obtain a high yield of the product [17]. Different effective statistical methods can be used to optimize enzyme production by screening the media composition that affects protease production significantly [11]. In this study, the production and optimization of alkaline protease were carried out by the Plackett–Burman (PB) design to screen various media components which yield maximum production of the enzyme. Furthermore, a multi-factorial response surface methodology (RSM) was used to study the effect of primary factors and their interactions on extracellular alkaline protease produced from Bacillus sp. [2]. Furthermore, protease enzyme characterization and kinetic studies were carried out from the isolate Bacillus cereus PW3A along with some industrial applications.
2. MATERIALS AND METHODS
2.1. Chemicals, Media, and Media Components
All the media components and chemicals were purchased from Sigma-Aldrich Chemicals Ltd (Missouri, USA), Ranbaxy Fine Chemicals Ltd (Mumbai, India), and HiMedia Laboratories Ltd (Mumbai, India)
2.2. Submerged Fermentation Production of the Enzyme
Isolation, identification, and characterization of B. cereus PW3A were reported in our earlier studies [18]. Screening of the microorganism for protease production was done on skimmed milk agar (SKM) plates as reported earlier [19]. Protease production was done using a production medium containing (g/100 ml): 1% glucose, 0.5% casein, 0.55% yeast extract, 0.2% KH2PO4, 1% Na2CO3 and 0.2% MgSO4.7H2O, and pH 8.0, while shaking at 120 rpm. Protease activity was assayed periodically and the isolates with the highest yield, B. cereus PW3A was selected for further studies. The microorganism B. cereus PW3A was transferred from their slant cultures into 250 ml conical flasks containing 100 ml nutrient broth. The cultures were grown at 37°C on a rotary shaker at 200 rpm for 24 h which was used as the inoculum for the submerged fermentation production of protease enzyme. From the prepared inoculum, 2 ml was added into 250 ml conical flasks containing 100 ml of production media. The production media were incubated at 37°C, at pH 8.0 for an incubation period of 48 h. After the incubation period, the cultured medium was centrifuged at 7000 rpm for 15 min at 4°C and the cell-free supernatant was used as the crude enzyme.
2.3. Determination of Alkaline Protease Activity
Protease activity (caseinolytic) was assayed by a modified method of Kunitz [20]. The enzyme reaction sample (1.0 ml) consisted of 500 L of 1% alkali-soluble casein in Tris-HCl buffer (pH 8.0) and 500 L of enzyme solution or cell-free supernatant. The enzyme reaction was started by adding an enzyme solution at 37°C. The solution was mixed by swirling and incubated at room temperature for 15 min. To terminate the reaction, 10% trichloroacetic acid (TCA, 5 ml) was added. The reaction mixture was incubated on ice for another 10 min. Then, the reaction mixture was subjected to centrifugation at 10,000 rpm for 10 min at 4°C. To 1.0 ml of supernatant, 5.0 ml of a 0.4 M Na2CO3 solution and 1.0 ml of Folin–Ciocalteu reagent were added and incubated for 30 min at 40°C. The L-tyrosine released from digested casein was quantified using a UV-Visible spectrophotometer at 660 nm. The standard curve was plotted using L-tyrosine as a standard. One unit of enzyme activity was expressed as the quantity of protease that releases 1 M/ml of tyrosine per minute. The enzyme activity was calculated by the formula given below:
Protein quantification of the sample was estimated based on Bradford’s method [21] using Bradford’s reagent (Sigma, USA).
2.4. Carbon Content Estimation
The carbon content was determined using a glucose standard graph. The estimation of carbon content was carried out using the DNS method where 1 ml of 3,5-dinitrosalicylic acid (DNS) was added to 1 ml of the crude enzyme and incubated for 10 min in a boiling water bath [22]. Subsequently, 8 ml of distilled water was added and the absorbance values were measured at 540 nm. Profiling studies were performed by comparing the production of protease enzyme to the change in carbon content and the change in biomass using suitable assays. A graph was plotted for protease activity versus carbon content to study the growth characteristics of the microorganism under study.
2.5. Optimization Studies for Media Ingredients
2.5.1. OFAT analysis
A “OFAT” approach was applied to understand the impact of different conditions on protease production and to examine and identify the important factors. Hence, in each experiment, one element at a time was changed while others were fixed. Various carbon sources (1% w/v) such as glucose, fructose, galactose, and lactose; various nitrogen sources (0.2% w/v) such as peptone, yeast extract, urea, and beef extract; a casein substrate (0–2% w/v) was used. To evaluate the combined effects of variables and optimum conditions for protease production, the PB design and RSM were used [23]. For optimization studies, the carbon and nitrogen source which exhibited the highest enzyme activity was selected.
2.5.2. PB design
Screening of different variables affecting protease production was performed by PB design [23]. In this study, two levels of six independent variables (fructose, yeast extract, casein, KH2PO4, Na2CO3, and MgSO4 7H2O) were evaluated as shown in Table 1. These variables were selected for a twelve-run PB design, and the effect of the individual variable was determined by the following equation.
E (xi) = 2 (qMi+- Mi-)/N
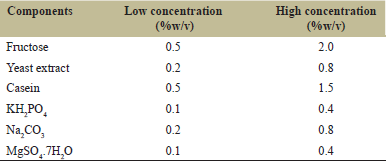 | Table 1: Components of production medium for protease production with their high and low ranges. [Click here to view] |
Where, E (xi), denotes the concentration effect of the test variable, Mi+ and Mi- are the total enzyme produced from the trials where the measured variable (xi) was calculated at high and low concentrations, respectively, and N signifies the number of trials [6].
According to the design matrix, 11 different combinations of significant variables were selected according to the protease activity measured. Minitab16 was used to generate and analyze the PB experimental design. The experiment was performed in triplicate. The PB design is on the first-order model given by the following equation:
Y= β0 + Σβi xi
Where Y, β0, βi, i, and xi are the dependent variable (response in terms of enzymatic activity), the intercept model, the estimated coefficient of the variable, the number of variables, and the independent variable, respectively. F-test is used for the analysis of variance (ANOVA) to define the meaning of each term in the equation and the statistical significance of the model. To evaluate the statistical significance of the regression coefficients, the student’s t-test was used. The competence of the model can be assessed by the lack of multiple coefficients of determination (R2) and its value. Variables with P-values (>0.1) are considered to have a significant impact on protease productivity.
2.5.3. RSM
Two independent variables, namely, fructose and yeast extract were optimized for increased production of protease enzyme by B. cereus PW3A using RSM [24,25]. Minitab 16 software (USA) was used to predict 11 experimental runs. The average value of three trials of the experiment was taken as a response value (Y). The main effect of the parameters was calculated at three levels (−1, 0, and +1) on protease production was analyzed. The central coded value of the variables was considered as zero. The investigated parameters ranging from low to high along with actual and predicted levels of the complete experimental plan are presented in Table 2. To understand the significance of the generated model from RSM, analysis of variance data was obtained. The “main effect” of all the variables on protease production was given by the Pareto chart. The significant effects (P < 0.5) were obtained by the Pareto chart and statistical analysis. These factors were included and varied for optimization studies using RSM. The response generated to the effect of variables in RSM was calculated by the following second-order polynomial equation.
Y= βo + β1X1 + β2 X2 + β11 X12 + β22X22 + β12X1 X2
Where, Y denotes the response generated, βo refers to the constant, β1, β2 and β11, β22 are the coefficients of linear terms and quadratic terms, respectively. β12 refers to the coefficient of the cross-product term. X1 and X2 signify the factors, fructose, and yeast extract, respectively.
2.6. Purification of Protease Enzyme
A two-step strategy, ammonium sulfate precipitation and dialysis were adopted for purification of protease from B. cereus PW3A. The crude protease was subjected to ammonium sulfate precipitation at various percentages of saturation levels (10–80%). The precipitated fraction was collected by centrifugation and dissolved in an appropriate amount of Tris HCl buffer (50 mM, pH 9.0) followed by dialysis. The protein sample obtained was checked for their enzyme activity and protein content. The specific activity of the protein was expressed as units per mg protein. The partially purified enzyme was used for further enzyme characterization.
 | Table 2: Various levels of factors for Response Surface Methodology. [Click here to view] |
2.7. Enzyme Characterization: Influence of Temperature, pH and Metal Ions on Protease Activity
2.7.1. Influence of temperature on enzyme activity
The thermostability of the enzyme was studied at a temperature range of 20–80°C. Partially purified enzymes were subjected to protease assay at different temperatures such as 20, 30, 40, 50, 60, 70, and 80°C.
2.7.2. Influence of pH on enzyme activity
The effect of pH (pH range 4.0–10.0) on enzyme activity was analyzed by conducting a protease assay on the partially purified enzyme samples.
2.7.3. Influence of metal ions on enzyme activity
Dialyzed enzyme was subjected to protease activity where the enzyme was pre-incubated in the presence of different metal ions (Na+, Cu+, Hg+, and Mg) at different concentrations (1 mM, 5 mM, and 10 mM).
2.7.4. Kinetic properties of protease
The kinetic parameters of partially purified enzyme were determined using various concentrations of casein (0.2%, 0.4%, 0.6%, 0.8%, 1.0%, 1.2%, 1.4%, 1.6%, 1.8%, and 2.0%) with standard assay conditions. The kinetic rate constants Km and Vmax were estimated using the Michaeli–Menten equation and Lineweaver Burk plot [26].
2.8. Statistical Analysis
All the data reported in the current study represent an average value obtained from triplicates from the experimental results and standard deviation was calculated.
3. RESULTS AND DISCUSSION
3.1. Microorganism and Culture Conditions
In our previous study, the extracellular protease-producing bacteria were isolated from a polluted water sample collected from a treatment plant at Hubballi City. Phenotypic studies indicated the isolate as circular, off-white, smooth with a large colony. Microscopically, the microbe appeared as spore-forming, rod-shaped, and Gram-positive bacillus. The isolated strain has shown optimal growth over a wide range of temperatures (15–45°C). The 16S rDNA gene sequencing analysis showed that this strain had the highest homology (98%) with B. cereus (GenBank ID: KT808623) and hence was identified as B. cereus PW3A as reported in our earlier studies [18]. The strain demonstrated an effective zone of clearance around its colony on skim milk agar indicating its ability to produce protease enzyme [Figure 1]. In terms of industrial applications, the selection of strain with a higher production rate of alkaline protease is important in submerged fermentation.
3.2. Time Course Profile of Growth and Extracellular Protease Production
The carbon content estimation was carried out by the DNS method. A modified Kunitz method was used to assay the enzyme activity of the protease. The results obtained were used to generate data for a profiling study which was conducted by comparing carbon estimation studies and protease activity to the change in biomass with time as represented in Figures 2 and 3. Protease activity increased with an increase in the growth of the organism [Figure 2]. Meanwhile, the glucose content shows a decreasing trend as the biomass concentration increases along the time of incubation [Figure 3]. The highest protease activity (17.229 U/ml) was observed after 48 h of incubation. It is also seen that after 60 h of incubation, the protease activity was observed to be decreasing. Similar results of optimum production after 48 h were reported by earlier studies [7,16,27]. The glucose content in the media follows a declining trend as the incubation period progresses while the enzyme activity increases. As the incubation time increases, the microbial growth also increases due to the consumption of carbon sources for energy production and growth. This energy is used to break down the substrate aiding in the increase in enzyme activity. After reaching a certain point, the cells enter a stationary phase establishing equilibrium between new cells and the dying cells. The increase in the biomass in the medium results in low optical density reading and hence the declining trend.
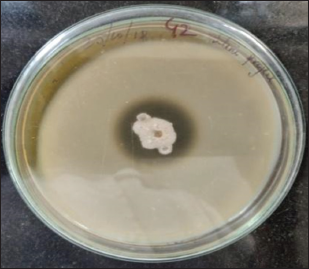 | Figure 1: Proteolytic activity demonstrated by Bacillus cereus PW3A. Zone of Clearance indicates hydrolysis of the substrate casein. [Click here to view] |
 | Figure 2: Profiling studies for growth and enzyme production from Bacillus cereus PW3A where the secondary Y-axis represents optical density (OD) values. [Click here to view] |
 | Figure 3: Profiling studies for growth and utilization of carbon source from Bacillus cereus PW3A where the secondary Y-axis represents optical density (OD) values. [Click here to view] |
3.3. Effect of Carbon and Nitrogen Source
To study the effect of various carbon sources on enzyme activity, glucose, fructose, galactose, and lactose was incorporated in the production media. Each carbon source was added at a level of 1% (w/v) in the inoculum and incubated. After 48 h, the sample was withdrawn and enzyme assay was performed as defined earlier. Figure 4 shows the effect of different carbon sources on the enzyme activity, the highest enzyme activity was observed in the presence of fructose resulted in 19.476 U/ml in comparison to glucose (16.858 U/ml), galactose (12.927 U/ml), and lactose (12.46 U/ml). Similarly, to study the effect of various nitrogen sources: Yeast extract, beef extract, peptone, and urea were incorporated at a level of (0.2%) keeping 0.5% glucose as carbon source. The result is represented in Figure 5. Among all the nitrogen sources used, yeast extract gave the highest enzyme activity with 17.269 U/ml and beef extract, Peptone and urea gave comparatively less enzyme activity 11.115 U/ml, 13.073 U/ml, and 8.427 U/ml, respectively. Yeast extract fructose was selected as nitrogen sources and carbon, respectively, for further studies as they have shown a significant effect on protease activity.
The higher production of alkaline protease depends on the media sources such as nitrogen and carbon [28]. Although B. cereus PW3A was capable to use a variety of carbon sources, the protease activity was different with each carbon source [Figure 4]. The fructose resulted as the optimal carbon source for alkaline protease production by B. cereus PW3A. Glucose and galactose provided a moderate to significant protease activity. The result is consistent with the previous studies using fructose as a carbon source to increase enzyme production in Bacillus sp [24,29]. Among different nitrogen sources tested by OFAT method, the optimum nitrogen source for maximum protease production was found to be yeast extract [Figure 5]. The results are in line with the previous findings on Bacillus sp. [30]. The nitrogen requirement for protease production differs from organism to organism. The media containing nitrogen and carbon sources affect the production of alkaline proteases [4].
 | Figure 4: Effect of various carbon sources on protease production. [Click here to view] |
 | Figure 5: Effect of various nitrogen sources. [Click here to view] |
3.4. Optimization Study: PB Design and RSM
PB design was used to determine the variables which had a significant effect on the protease enzyme activity. The significant variables were needed to be optimized to have an optimal composition of the medium for maximum enzyme activity. Variables such as fructose and yeast extract were confirmed from PB design experiments and RSM was used to predict optimum levels of the variables. PB design was used to develop a matrix containing 11 runs with the combinations of the high and low values of all the components in the medium as shown in Table 1. The significant factors were identified based on the estimated effects and regression analysis. The analysis of the optimization study showed that two factors were significant as reported by their P values. Factors having P < 0.05 are considered to have a significant effect on the response. The Pareto chart obtained from the PB experiment suggested similar results as obtained from the statistical data [Figure 6]. The P-values of fructose and yeast extract were 0.033 and 0.018, respectively, which are <0.05, demonstrating their importance in the media composition. Table 3 gives the estimated coefficient of each component in the media. The values derived from the student’s t-test and P-value are used to understand the importance of the coefficients. The larger the extent of t-value and smaller P-value, the more significant is the corresponding coefficient. The values of fructose and yeast extract coincide with the requirements of the significant coefficient. This data of variables were evaluated for further optimization.
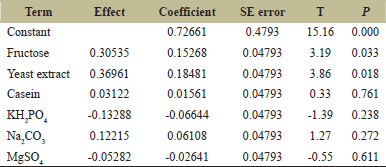 | Table 3: Estimated Effects and coefficients for enzyme activity (PB Design). [Click here to view] |
 | Figure 6: Pareto chart for screening significant variables for protease production where enzyme activity (EA) is the response. [Click here to view] |
Three levels of fructose and yeast extract were tested using this experiment as shown in Table 2. Further Table 4 shows details of the experimental design for the PB experiment. The matrix developed gave a set of 13 experiments with different combinations of fructose and yeast extract levels as shown in Table 5. Estimated effects and coefficients for enzyme activity derived from PB design are shown in Table 3. The response from the RSM experiment was studied using the analysis of variance (ANOVA). The combination of medium components in run 2 gives the highest activity of 47.43 U/ml of the protease enzyme. The statistical results obtained are shown in Table 6. The regression equation was obtained from the analysis of variance (ANOVA) which indicated that the R2 value for the experiment is 0.6309 (a value of R2 > 0.75 indicates the aptness of the model). The R2 is the proportion of variability in response values explained or accounted for by the model; it can explain 63.09% variation in the response. The results of RSM were also interpreted in the 3D response surface curves [Figure 7] and their respective 2D contour plots [Figure 8] which was used to determine the optimum levels of significant variables for maximum response. These plots were responsible for providing a visual interpretation of the interaction between fructose and yeast extract. Maximum activity was observed to be 47.43 U/ml when the concentration of fructose is high and that of yeast extract is low. Figure 8 provides the visual representation of the same and reflects the negative effect of high yeast extract concentration on protease production. These results are in line with the data obtained on protease production by B. clausii [9]. The reason for the negative effect of high yeast extract concentration was reported to be the growth-independent repression of protease production [9]. Some studies show that repressive roles are played by organic nitrogen sources and excessive amino acid and ammonium ions in alkaline protease production [28]. An overall 2.7-fold increase in protease production was achieved after optimization by using RSM. Thus, it is evident that the statistical approach considers the interaction of independent variables to provide a basis for the model of experiments which shows the nonlinear nature of the response in a short-term experiment [31]. The acceptance rate of the statistical experimental design by researchers is growing in the biotechnology sector due to the satisfactory optimization of protease production from microbial sources using statistical approaches. Using statistical design to screen and optimize culture conditions allows quick identification of the important factors and interactions between them [32].
 | Figure 7: 3D response surface plots depicting the interaction of yeast extract and fructose for protease production where enzyme activity (EA) is the yield. [Click here to view] |
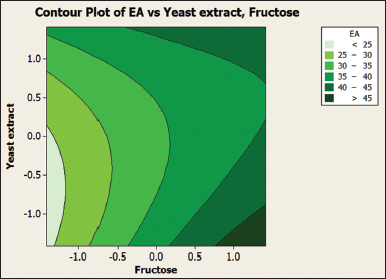 | Figure 8: 2D contour plots for studying the interaction of yeast extract and fructose for maximum enzyme activity (EA). [Click here to view] |
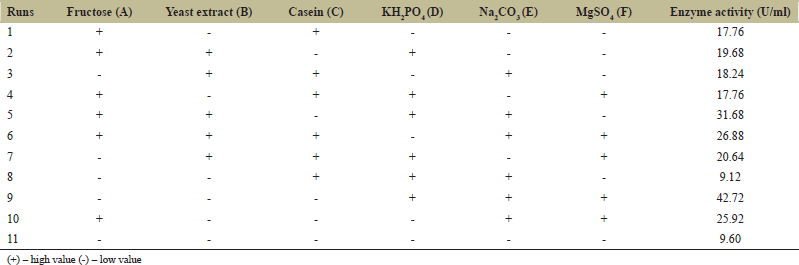 | Table 4: Design matrix of different combinations for the media components for Plackett–Burman design. [Click here to view] |
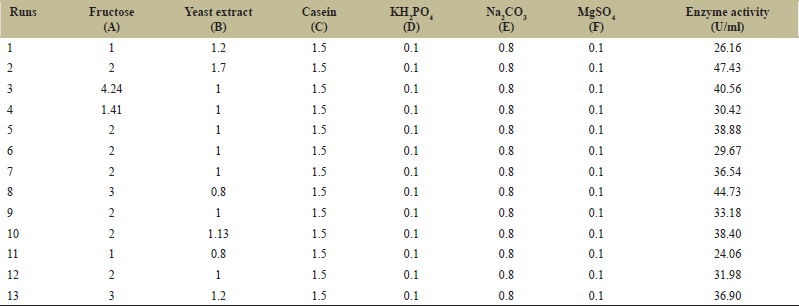 | Table 5: Design matrix of different combinations for the media components for Response Surface Methodology. [Click here to view] |
 | Table 6: Analysis of Variance for Response Surface Methodology (RSM). [Click here to view] |
3.5. Partial Purification of Protease
After 48 h of incubation, the cell-free supernatant was used for the partial purification (saturation level, 65% w/v) of the alkaline protease produced by B. cereus strain PW3A. The protein precipitate obtained was dissolved and dialyzed overnight against the phosphate buffer. Table 7 shows the results of enzyme purification by ammonium sulfate precipitation and dialysis. The partially purified enzyme is further used for characterization studies.
 | Table 7: Purification summary of alkaline protease from Bacillus cereus PW3A. [Click here to view] |
3.6. Enzyme Characterization: Influence of Temperature, pH, and Metal Ions on Protease Activity
3.6.1. Effect of temperature and pH on protease activity
Figure 9 shows the effect of temperature on enzyme activity with a range from 20°C to 80°C. It is evident from the figure that at 50°C the enzyme activity was highest (18.7125 U/ml). Hence, the optimum temperature was recorded to be 50°C. However, it is also observed the enzyme retained significant activity between 40°C and 60°C indicating its thermal stability. The effect of pH on enzyme activity was studied at a pH range from 4 to 10. The highest enzyme activity was reported at pH 8, with an activity of 17.36 U/ml. The enzyme also showed stability with significant enzyme activity at a range of pH 8 to pH 10 indicating its alkaline nature with 8 being the optimum pH [Figure 10].
Temperature and pH requirements are extremely important as they are considered among the factors that affect the growth and production of metabolites by microorganisms [29,33]. There is an optimum temperature and pH for each enzyme which gives the maximum enzyme activity. In the present study, however, the most significant level of growth and production of protease was supported by temperature ranging between 50 and 55?C [Figure 9] with a rapid decline after 50°C. Bacillus sp. APP1 showed identical results to the optimum temperature for enzyme production as documented [34]. It was reported that the optimum temperature for alkaline protease by Bacillus stearothermophilus AP-4 was found to be 55°C [35]. Protein conformation remains stable, but at a narrow range of increasing temperature. The protein destabilizes because of abrupt loss of structure when weak interactions like hydrogen bonds are affected [33]. Despite the reduced activity of the protease enzyme when the temperature increases, it can be established as a thermo-stable enzyme as observed from the graphs and find applications in industries with elevated temperatures.
The optimum pH for the activity of the protease enzyme produced by B. cereus PW3A was 8.0. The results were comparable with the results obtained from studies conducted on the characterization Bacillus sp. CA15 [36] showed optimum activity at pH 8- 8.5. It was also reported that the optimum protease activity was obtained at pH 8.5 for B. subtilis isolate [10]. There has been evidence that the optimum pH for alkaline proteases of Bacillus sp. varies from 8 to 11 and they are species-specific [15]. Protease activity was observed at pH 8 according to the study performed on Bacillus licheniformis [37]. At low and high pH, the ionization states of amino acid changes play a crucial role in interactions that maintain the three-dimensional structure of the protein or the enzyme [36]. The increase or decrease in the hydrogen ion concentration in the reaction mixture changes the native conformation and results in enzyme denaturation. The present study helps to establish the stability of the protease enzyme by B. cereus PW3A that showed notable enzyme activity at pH 8–pH 10 (optimum enzyme activity was at pH 8 indicating its alkaline nature, Figure 10). The enzyme will find applications in a wide variety of large and small-scale activities since it is stable at a wide range of pH.
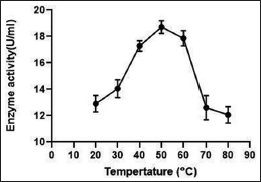 | Figure 9: Effect of temperature on protease activity produced from Bacillus cereus PW3A. [Click here to view] |
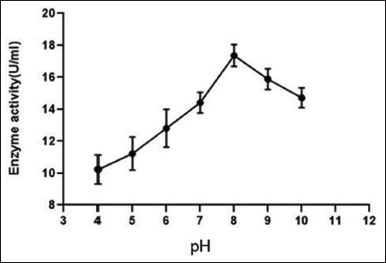 | Figure 10: Effect of pH on protease activity produced from Bacillus cereus PW3A. [Click here to view] |
3.6.2. Effect of metal ions on enzyme activity
A protease assay was conducted with three different concentration levels of metal ions (1 mM, 5 mM, and 10 mM) to analyze its effect on the activity of the enzyme. Mg2+ and Cu2+ increased the enzyme activity by acting as activators while the Hg2+ demonstrated inhibitory function according to the results of the present study [Figure 11]. Na+ showed a negligible effect on enzyme activity compared to other metal ions, indicating that it does not have a profound effect on the enzyme. It was reported that Mo2+, Cu2+, Fe2+, Mg2+, Ni2+, and Ca2+ affected the enzyme activity positively of B. cereus S8 [5]. Mercury is a heavy metal and at a molecular level, it forms bonds with sulfhydryl groups on an enzyme. The binding of mercury can change the shape of the enzyme and block its activity.
3.6.3. Enzyme kinetic studies
The effect of casein concentrations on the enzyme activity was evaluated using different concentrations of casein with standard assay conditions. The enzyme-substrate curve (Michaelis- Menten graph) equation and Lineweaver Burk plot were used to determine the value of Km and Vmax. A lower Km value indicates a strong binding affinity of protease to the substrate. The accurate values of Km and Vmax from the enzyme-substrate curve (Michaelis–Menten graph) were unsatisfactory; therefore, the Lineweaver Burk plot [Figure 12] was used. The Km (0.6667 μM) and Vmax (15.873 U/ml) values were calculated from the Lineweaver Burk plot. For maximum activity, the optimal substrate (i.e., casein) concentration was determined in terms of Michaelis–Menten kinetic constant (Km) and the maximum velocity (Vmax). The Vmax denotes the maximum enzymatic rate of reaction where the enzyme is saturated with substrate and the affinity of enzyme and with its substrate is expressed by Km [38]. An extracellular alkaline protease from a novel haloalkaliphilic bacterium reported a Km and Vmax of protease of 0.153 g/100 ml and 454 U/ml, respectively [39]. The Km of the alkaline protease from B. licheniformis Bl8 with the substrate, casein was found to be 3.2 mg/ml [40].
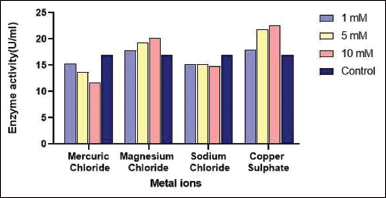 | Figure 11: Effect of metal ions on protease activity produced from Bacillus cereus PW3A. [Click here to view] |
 | Figure 12: Lineweaver Burk plot for partially purified protease from Bacillus cereus PW3A. [Click here to view] |
The current research investigated the studies on optimization of media components for the production of protease enzyme from B. cereus PW3A using statistical and non-statistical approaches, parameter characterization (temperature, pH, and effects of metal ions), kinetic studies, and purification of protease enzyme. This combination of studies is less explored and reported in an individual paper. Stability is a very important property of industrial enzymes. The protease produced by B. cereus PW3A has its optimum temperature of 50ºC and hence it is advantageous that it helps contribute to the reduction of microbial contaminants as well as cooling costs. The enzyme does not get denatured after the optimum pH (pH 8) and is stable up to pH 10 and hence finds applicability in industrial processes requiring an alkaline pH range of 8–10 thus making it an economical source of protease enzyme.
4. CONCLUSION
The present study demonstrated how optimizing the media composition and studying different parameters can affect enzyme activity. Statistical experimental approaches proved crucial in optimizing the production of protease by B. cereus PW3A, which resulted in a 3-fold increase in enzyme activity. The enzyme exhibited temperature and pH stability that may be useful in the process of having alkaline environments and with a wide range of temperatures. The study also demonstrated that magnesium and copper acted as activators and mercury played a role as an inhibitor for protease production from B. cereus. Further investigation can be done for process parameters pH, temperature, agitation, speed, dissolved oxygen, etc., which affect enzyme production.
5. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis, and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
6. ACKNOWLEDGMENT
The authors acknowledge KLE Technological University for providing laboratory facilities.
7. FUNDING
The study is supported by Capacity Building Fund from KLE Technological University.
8. CONFLICT OF INTEREST
The authors report no financial or any other conflicts of interests in this work.
9. ETHICAL APPROVALS
The study does not involve any animals or human subjects.
10. DATA AVAILABILITY
All the required data is provided in the manuscript.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Jaouadi NZ, Jaouadi B, Hlima HB, Rekik H, Belhoul M, Hmidi M, et al. Probing the crucial role of Leu31 and Thr33 of the Bacillus pumilus CBS alkaline protease in substrate recognition and enzymatic depilation of animal hide. PLoS One 2014;9:e108367. CrossRef
2. Sathishkumar R, Ananthan G, Arun J. Production, purification and characterization of alkaline protease by ascidian associated Bacillus subtilis GA CAS8 using agricultural wastes. Biocatal Agric Biotechnol 2015;4:214-20. CrossRef
3. Naveed M, Nadeem F, Mehmood T, Bilal M, Anwar Z, Amjad F. Protease-a versatile and ecofriendly biocatalyst with multi-industrial applications: An updated review. Catal Lett 2021;151:307-23. CrossRef
4. Mothe T, Sultanpuram VR. Production, purification and characterization of a thermotolerant alkaline serine protease from a novel species Bacillus caseinilyticus. 3 Biotech 2016;6:53. CrossRef
5. Lakshmi BK, Kumar DM, Hemalatha KP. Purification and characterization of alkaline protease with novel properties from Bacillus cereus strain S8. J Genet Eng Biotechnol 2018;16:295-304. CrossRef
6. Saxena R, Singh R. Statistical optimization of conditions for protease production from Bacillus sp. Acta Biol Szeged 2010;54:135-41.
7. Bajaj BK, Jamwal G. Thermostable alkaline protease production from Bacillus pumilus D-6 by using agro-residues as substrates. Adv Enzyme Res 2013;1:30-6. CrossRef
8. Joo HS, Kumar CG, Park GC, Paik SR, Chang CS. Oxidant and SDS?stable alkaline protease from Bacillus clausii I?52: Production and some properties. J Appl Microbiol 2003;95:267-72. CrossRef
9. Oskouie SF, Tabandeh F, Yakhchali B, Eftekhar F. Response surface optimization of medium composition for alkaline protease production by Bacillus clausii. Biochem Eng J 2008;39:37-42. CrossRef
10. Hameed A, Natt MA, Evans CS. Production of alkaline protease by a new Bacillus subtilis isolate for use as a bating enzyme in leather treatment. World J. Microbiol Biotechnol 1996;12:289-91. CrossRef
11. Liu S, Fang Y, Lv M, Wang S, Chen L. Optimization of the production of organic solvent-stable protease by Bacillus sphaericus DS11 with response surface methodology. Bioresour Technol 2010;101:7924-9. CrossRef
12. Shivanand P, Jayaraman G. Production of extracellular protease from halotolerant bacterium, Bacillus aquimaris strain VITP4 isolated from Kumta coast. Process Biochem 2009;44:1088-94. CrossRef
13. Sen S, Veeranki VD, Mandal B. Effect of physical parameters, carbon and nitrogen sources on the production of alkaline protease from a newly isolated Bacillus pseudofirmus SVB1. Ann Microbiol 2009;59:531-8. CrossRef
14. Rao CS, Sathish T, Ravichandra P, Prakasham RS. Characterization of thermo-and detergent stable serine protease from isolated Bacillus circulans and evaluation of eco-friendly applications. Process Biochem 2009;44:262-8. CrossRef
15. Guangrong H, Tiejing Y, Po H, Jiaxing J. Purification and characterization of a protease from thermophilic Bacillus strain HS08. Afr J Biotechnol 2006;5:2433-8.
16. Asha B, Palaniswamy M. Optimization of alkaline protease production by Bacillus cereus FT 1 isolated from soil. J Appl Pharm Sci 2018;8:119-27.
17. Adetunji AI, Olaniran AO. Statistical modelling and optimization of protease production by an autochthonous Bacillus aryabhattai Ab15-ES: A response surface methodology approach. Biocatal Agric Biotechnol 2020;24:101528. CrossRef
18. Babruwad PR, Prabhu SU, Upadhyaya KP, Hungund BS. Production and characterization of thermostable polyhydroxybutyrate from Bacillus cereus PW3A. J Biochem Technol 2016;6:990-5.
19. Bajaj BK, Sharma P. An alkali-thermotolerant extracellular protease from a newly isolated Streptomyces sp. DP2. N Biotechnol 2011;28:725-32. CrossRef
20. Amoozegar MA, Fatemi AZ, Karbalaei-Heidari HR, Razavi MR. Production of an extracellular alkaline metalloprotease from a newly isolated, moderately halophile, Salinivibrio sp. strain AF-2004. Microbiol Res 2007;162:369-77. CrossRef
21. Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248-54. CrossRef
22. Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 1959;31:426-8. CrossRef
23. Plackett RL, Burman JP. The design of optimum multifactorial experiments. Biometrika 1946;33:305-25. CrossRef
24. Kumar RS, Ananthan G, Prabhu AS. Optimization of medium composition for alkaline protease production by Marinobacter sp. GA CAS9 using response surface methodology-a statistical approach. Biocatal Agric Biotechnol 2014;3:191-7. CrossRef
25. Hegde S, Bhadri G, Narsapur K, Koppal S, Oswal P, Turmuri N, et al. Statistical optimization of medium components by response surface methodology for enhanced production of bacterial cellulose by Gluconacetobacter persimmonis. J Bioprocess Biotech 2013;4:1-5.
26. Parthasarathy M, Gnanadoss JJ. Purification and characterization of extracellular alkaline protease from Streptomyces sp. LCJ12A isolated from Pichavaram mangroves. J Appl Boil Biotechnol 2020;8:15-20. CrossRef
27. Olajuyigbe FM, Ehiosun KI. Production of thermostable and organic solvent-tolerant alkaline protease from Bacillus coagulans PSB-07 under different submerged fermentation conditions. Afr J Biotechnol 2013;12:3341-50.
28. Gupta R, Beg Q, Khan S, Chauhan B. An overview on fermentation, downstream processing and properties of microbial alkaline proteases. Appl Microbiol Biotechnol 2002;60:381-95. CrossRef
29. Cheng SW, Wang YF, Wang ML. Statistical optimization of medium compositions for alkaline protease production by newly isolated Bacillus amyloliquefaciens. Chem Biochem Eng Q 2012;26:225-31.
30. Shine K, Kanimozhi K, Panneerselvam A, Muthukumar C, Thajuddin N. Production and optimization of alkaline protease by Bacillus cereus RS3 isolated from desert soil. Int J Adv Res Biol Sci 2016;3:193-202.
31. Puri S, Beg QK, Gupta R. Optimization of alkaline protease production from Bacillus sp. by response surface methodology. Curr Microbiol 2002;44:286-90. CrossRef
32. Chauhan B, Gupta R. Application of statistical experimental design for optimization of alkaline protease production from Bacillus sp. RGR-14. Process Biochem 2004;39:2115-22. CrossRef
33. Desai SS, Hungund BS. Submerged fermentation, purification, and characterization of L-asparaginase from Streptomyces sp. isolated from soil. J Appl Biol Biotechnol 2018;6:17-23. CrossRef
34. Chu WH. Optimization of extracellular alkaline protease production from species of Bacillus. J Ind Microbiol Biotechnol 2007;34:241-5. CrossRef
35. Dhandapani R, Vijayaragavan R. Production of a thermophilic, extracellular alkaline protease by Bacillus stearothermophilus AP-4. World J. Microbiol Biotechnol 1994;10:33-5. CrossRef
36. Uyar F, Porsuk I, Kizil G, Yilmaz EI. Optimal conditions for production of extracellular protease from newly isolated Bacillus cereus strain CA15. Eurasian J Biosci 2011;5:1-9. CrossRef
37. Suganthi C, Mageswari A, Karthikeyan S, Anbalagan M, Sivakumar A, Gothandam KM. Screening and optimization of protease production from a halotolerant Bacillus licheniformis isolated from saltern sediments. J Genet Eng Biotechnol 2013;11:47-52. CrossRef
38. Rehman HU, Aman A, Silipo A, Qader SA, Molinaro A, Ansari A. Degradation of complex carbohydrate: Immobilization of pectinase from Bacillus licheniformis KIBGE-IB21 using calcium alginate as a support. Food Chem 2013;139:1081-6. CrossRef
39. Patel RK, Dodia MS, Joshi RH, Singh SP. Purification and characterization of alkaline protease from a newly isolated haloalkaliphilic Bacillus sp. Process Biochem 2006;41:2002-9. CrossRef
40. Lakshmi G, Prasad NN. Purification and characterization of alkaline protease from a mutant Bacillus licheniformis Bl8. Adv Biol Res 2015;9:15-23.