1. INTRODUCTION
Hepatitis C is a liver disease caused through infection by the Hepatitis C virus (HCV). It is an inflammation of the liver, observed only in humans and chimpanzees. Broadly, hepatitis is caused by viral strain, namely, Hepatitis A, B, C, D, and E [1,2]. Out of these five viruses, Hepatitis B, C, and D cause acute and chronic hepatitis. HCV is a small, enveloped, single-stranded RNA virus belonging to the family Flaviviridae and the genus Hepacivirus. The family Flaviviridae includes yellow fever, West Nile, and dengue viruses [3]. HCV is a blood-borne pathogen and a major cause of liver diseases, causing acute and chronic hepatitis [2]. If left untreated, it will likely progress to cirrhosis and hepatocellular carcinoma. Early acute infection is mostly asymptomatic. Hence, around 50% of HCV people do not realize that they are infected. This allows the progression of infection, and it is likely that Hepatitis C is the leading cause of liver necrosis and liver cancer [4].
Approximately 120–130 million people are infected by HCV worldwide, of which 58 million develop chronic hepatitis. Annually, there are 1.5 million new infections, and in 2019, approximately 290,000 people die due to Hepatitis C. In the USA, Hepatitis C has a higher mortality rate than human immunodeficiency virus 1. Hepatitis C infection is treated with antiviral medicine such as ribavirin solely or in combination with sofosburin and simeprevir. No effective vaccine has been developed against HCV due to its high variability in the viral genome [5].
HCV particles are spherical, and their size ranges from 40 to 80 nm. The structural elements of this typical virus include envelope glycoproteins, lipid membrane, nucleocapsid, and RNA genome. The process of the viral life cycle is initiated by the entry of HCV into hepatocytes [6]. Several ligand-receptor interactions mediate the attachment of virus to the hepatocytes. Initially, low-affinity attachment of the virus is believed to involve viral glycosaminoglycans and low-density lipid receptors. HCV consists of envelope proteins – E1 and E2- highly conserved and glycosylated. E2 contains C-terminal transmembrane domains that form a heterodimer, enabling viral entry into the host cells. Receptor-binding amino acid residues – Tyr527, Trp529, Gly530, and Asp535, are present in the C-terminal transmembrane domain [4]. The interactions between ligands and these amino acid residues are crucial for viral attachment. E1 is approximately 192 amino acids long, and E2 is about 363 amino acids long. HCV E2 glycoprotein is considered to interact with two hepatic receptors of host hepatocytes, i.e., scavenger receptor Class B type 1 and the tetraspanin cluster of differentiation (CD81) receptor.
E2-CD81 interaction is essential for viral adsorption onto hepatocytes. E1 does not interact directly with host receptors. It is believed to maintain a functional conformation of E2 necessary for binding to the cellular membrane receptors. HCV entry through clathrin-mediated endocytosis is followed by fusion of virus and endosomal membranes. E1 facilitates the fusion of the virus to the endosome, which is the final event of infection [3-10]. Ribavirin, which is the standard approved drug for the treatment of HCV, has been reported to have adverse effects such as anemia, arrhythmia, hypocalcemia, chest pain, dizziness, nausea, hyperuricemia and hyperbilirubinemia [11]. Thus, the study aims to identify a possible alternative medicine, preferably of a plant source, to treat Hepatitis.
The genus Nardostachys is endemic to the Himalayas, with immense commercial demand due to its medicinal properties. It is a perennial herb whose size ranges from 10 to 60 cm in height. The genus contains only one species, Nardostachys jatamansi. The plant is commonly known as jatamansi or spikenard, which grows best in steep, undisturbed grassy, rocky, and sandy loamy soil [12]. N. jatamansi has been reportedly used to treat various disorders, such as digestive, nervous, respiratory, urinary, circulatory and reproductive disorders [13]. It has also been reportedly used to cure cough, fever, headache, food poisoning, intestinal worms, joint pain, and to enhance memory. The plant extract has anti-diabetic, anti-hyperglycemic, anti-inflammatory, anti-Parkinsons, anti-tumor, anti-cancer, and radioprotective properties. Sesquiterpenes and its derivatives are this plant’s major active phytochemical constituents [14]. N. jatamansi is listed as critically endangered in the IUCN Red List of Threatened Species and is highly susceptible to overharvesting due to its high commercial value. In India, N. jatamansi is found in Arunachal Pradesh, Sikkim, Uttarakhand, and Himachal Pradesh [15,16]. However, plant tissue culture techniques had optimized their propagation [17].
Bioinformatics and computational tools have been implemented mainly for analyzing the efficacy and safety of potential drugs. Molecular docking is used to position computer-generated 3-D conformation of small ligands into the binding sites of target receptors. Docking studies provide a highly effective approach for the preliminary analysis of potential new drugs and reduce the time, cost, and labor associated with in vivo and in vitro assays of drug evaluation [18]. It is an integral part of Computer-aided drug design (CADD). Molecular docking studies are employed to analyze and compare the strength of binding affinities between target receptors and ligands. The findings of the docking studies could provide essential insights for drug design. AutoDock Vina is one of the fastest, most accurate, and most widely used open-source software for molecular docking [19]. Recently, AutoDock has been employed in screening phytochemicals against SARS-CoV-19, targeting its ACE2 receptor and Mpro protease, to inhibit viral entry and viral replication, respectively [20].
Similarly, there have been other multiple docking studies against Covid-19 [21-23]. Apart from SARS-CoV-19, there have been docking studies on colorectal cancer [24]. Successful docking of phytochemicals from Andrographis paniculata against Marburg virus has also been reported [25].
In this study, molecular docking was carried out using AutoDock Vina tools, Discovery Studio v20.1.0.19295, and other molecular visualizing tools such as PyMol and Avogadro [26]. Swiss absorption, delivery, metabolism, and excretion (ADME) was employed to predict the pharmacokinetics and drug-likeliness of the phytochemicals. Parameters such as water solubility, gastrointestinal (GI) absorption, permeation of blood–brain barrier (BBB), and lipophilicity. Were taken into consideration. Toxicity studies were carried out using pkCSM. Mutagenicity, hepatotoxicity, and hERG inhibition of selected phytochemicals were also evaluated using pkCSM. Both SwissADME and pkCSM are free online web tools [27,28]. This study aimed to find the best bioactive molecules of N. jatamansi against the HCV by inhibiting the formation of the E2-CD81 complex, which is critical for viral attachment and subsequent entry of HCV into the hepatocytes.
2. MATERIALS AND METHODS
The following are the open-source software used for this study: (i) Avogadro, (ii) Discovery Studio visualizer, (iii) AutoDockTools-1.5.7, (iv) PyMol, (v) SwissADME, (vi) pkCSM, and (vii) LigPlot+ v.2.2.
2.1. Selection and Preparation of Ligands
The 3-D structures of the bioactive molecules of N. jatamansi were downloaded in structure data file format from PubChem database [https://pubchem.ncbi.nlm.nih.gov/]. These ligands were subjected to energy optimization or minimization using Avogadro software and were further prepared using AutoDock MGLTools. The ligands were then saved in Protein Data Bank, Partial Charge (Q) and Atom Type (T) (PDBQT) format. The standard reference drug used in this study is ribavirin, and its 3-D structure was also downloaded from the PubChem database and subjected to similar processing and preparation as the phytochemicals.
2.2. Selection and Preparation of Receptor
This study’s selected viral protein receptor was the HCV’s E2 receptor. It was chosen as it interacts with the CD81 receptor of liver cells. This interaction is mandatory for HCV entry into the hepatocytes [7]. The 3-D structure of E2 was downloaded from the RCSB PDB database [https://www.rcsb.org/]. The first step in preparation of receptors is the removal of heteroatoms and water molecules. The receptor was then saved in PDB format and processed using AutoDock Tools with which native receptor ligands were removed, and missing atoms were repaired. The addition of polar hydrogens, Kollman, and computing Gasteiger charges was performed using AutoDock tools, and charge deficits were also spread over the atoms [29]. The receptor was then saved in PDBQT format.
2.3. Prediction of Binding Site and Molecular Docking
The binding site is the portion of the receptors wherein the ligands bind and cause chemical interaction. The coordinates of the binding site of the E2 receptor were retrieved using the Discovery Studio visualizer. After removing heteroatoms, the coordinates of the binding sites of the receptor were generated from either PDB site records or the receptor cavity sites. The coordinates of the grid obtained is X=36.385347, Y=−67.500194, and Z=−46.260347.
In our study, docking was carried out using AutoDock Tools 1.5.7, wherein the phytochemicals of N. jatamansi were docked against the viral E2 receptor [7,19]. Simultaneously, the reference drug, ribavirin was also docked against the E2 receptor. Phytochemicals showing a higher negative value of binding energy (kcal/mol) than ribavirin were considered for further studies.
Cognate docking was performed to validate the docking protocol. Without altering the parameters, the ligands were redocked onto the receptor’s binding site. Redocking was performed twice-the first redocking was carried out using the same software, i.e., AutoDock Vina, while the second redocking used a different software, CB-Dock.
2.4. ADME Studies and BOILED-Egg Prediction
SwissADME is a free online tool employed to study molecular pharmacokinetics, physicochemical properties, and drug-likeliness of small molecules. The major aspects taken into consideration by the web tool are ADME. Other parameters such as the Lipinksi’s rule of 5, Abbot bioavailability score, solubility, and lipophilicity are also considered to support drug discovery [27,30]. The phytochemicals’ absorption and BBB penetration capacity were also analyzed using BOILED-Egg (Brain or IntestinaL EstimateD permeation method). The BOILED-Egg is a graphical model that predicts the capacity of GI absorption and permeability of the BBB of small molecules of interest [31]. The canonical SMILES of the phytochemicals were obtained from PubChem and copied onto the SwissADME platform. Otherwise, the structures of the phytochemicals can be drawn manually as supported by the tool. ADME studies have increased drug discovery efficiency by reducing the time taken for pharmacokinetic-related studies during clinical trials.
2.5. Toxicity Studies
Analysis of the toxicity of the phytochemicals indicates the safety of the selected molecules for use as therapeutic drugs. This was performed using an online in silico web tool, pkCSM. The different toxicity parameters considered by pkCSM are-hepatotoxicity, AMES toxicity, hERG I and hERG II inhibitor, maximum tolerated dose, skin sensitization, oral rat chronic toxicity (lowest observed adverse effect level [LOAEL]), oral rat acute toxicity (LD50), minnow toxicity and Tetrahymena pyriformis toxicity [28]. The canonical SMILES of the phytochemicals were copied onto the pkCSM clipboard, and the toxicity assay was carried out.
3. RESULTS
3.1. Molecular Docking Analysis
The idea of docking phytochemicals against the E2 receptor stems from the phytochemicals’ potential of inhibiting the receptor and obstructing viral attachment onto the hepatocytes. The phytochemicals of N. jatamansi were docked against the E2 receptor of HCV. The presence of phytochemicals with higher negative binding energy than standard drugs for treating HCV infection may provide suitable drug candidates as alternatives for pre-existing drugs. The viral E2 receptor was docked against the phytochemicals of N. jatamansi using AutoDock Vina. The 3-D structure of the E2 receptor and its binding site are illustrated in Figures 1 and 2, respectively. The reference drug, ribavirin, was also docked against the viral receptor. The binding energy of ribavirin with the E2 receptor was calculated to be −5.4 kcal/mol.
 | Figure 1: 3-D structure of viral E2 receptor. [Click here to view] |
 | Figure 2: Binding site of E2 receptor. [Click here to view] |
However, among the 25 phytochemical compounds docked against the viral receptor, 17 compounds showed higher negative values than the reference compound drug, i.e., ribavirin. As most of the phytochemicals showed higher negative binding energy than the reference drug, the compounds were subjected to ADME studies. The binding energies of the phytochemicals against E2 and ribavirin are shown in Table 1. Three phytochemicals were prioritized based on their binding energies and ADME studies-ursolic acid, nardostachysin, and α-gurjunene. The binding energies of three selected phytochemicals against E2 were −7.8 kcal/mol for ursolic acid, −6.8 kcal/mol for nardostachysin, and −5.7 kcal/mol for α-gurjunene.
Table 1: Docking scores of ribavirin and the phytochemicals of Nardostachys jatamansi against viral E2 receptor.
| Phytochemicals | Canonical SMILES | PubChem CID | Binding energy (kcal/mol) |
|---|---|---|---|
| Ribavirin (reference drug) | C1=NC(=NN1C2C(C(C(O2)CO)O)O)C(=O)N | 37542 | −5.4 |
| Ursolic acid | CC1CCC2(CCC3(C(=CCC4C3(CCC5C4(CCC(C5(C)C)O)C)C)C2C1C)C)C(=O)O | 64945 | −7.8 |
| Taraxerone | CC1(CCC2(CC=C3C4(CCC5C(C(=O)CCC5(C4CCC3(C2C1)C)C)(C)C)C)C)C | 92785 | −7.7 |
| Lupeol | CC(=C) C1CCC2(C1C3CCC4C5(CCC(C(C5CCC4(C3(CC2)C)C)(C)C)O)C)C | 259846 | −7.0 |
| Nardostachysin | CC1CCC2C1CC(=CC=C2C(=O)OCC3(C(CC4C3C(=O)OCC4=C)O)O)C(C)C | 10598736 | −6.8 |
| Oroselol | CC(C)(C1=CC2=C(O1)C=CC3=C2OC(=O)C=C3)O | 160600 | −6.6 |
| Jatamansinone | CC1(C(=O)CC2=C(O1)C=CC3=C2OC(=O)C=C3)C | 759294 | −6.6 |
| Seselin | CC1(C=CC2=C(O1)C=CC3=C2OC(=O)C=C3)C | 68229 | −6.1 |
| Jatamansinol | CC1(C(CC2=C(O1)C=CC3=C2OC(=O)C=C3)O)C | 600670 | −6.1 |
| Dihydrojatamansin | CCC (C) C(=O)OC1CC2=C(C=CC3=C2OC(=O)C=C3)OC1(C)C | 11002035 | −6.1 |
| Oroselone | CC(=C)C1=CC2=C(O1)C=CC3=C2OC(=O)C=C3 | 74477 | −5.9 |
| Jatamol B | CC(=C)C1CCC2(CC(CC(=C)C2C1)OC(=O)CC(C)(C)O)C | 101618011 | −5.9 |
| Nardosinone | CC1CCC=C2C1(C3C(CC2=O)OOC3(C)C)C | 168136 | −5.8 |
| Jatamol A | CC(=C)C1CCC2(CC(CC(=C)C2C1)O)C | 101618010 | −5.8 |
| α-Gurjunene | CC1CCC2C(C2(C)C)C3=C(CCC13)C | 15560276 | −5.7 |
| Nardoaristolone | CC1CC(=O)C=C2C1(C3C(C2=O)C3(C)C)C | 71583464 | −5.6 |
| Valeranol | CC1CCC=C2C1(CC(CC2)C(C)(C)O)C | 6429378 | −5.5 |
| Angelicin | C1=CC2=C(C=CO2)C3=C1C=CC(=O)O3 | 10658 | −5.5 |
| Jatamansone | CC(C)C1CCC2(CCCC(=O)C2(C1)C)C | 10198387 | −5.4 |
| Spirojatamol | CC(C)C1CCC(=C)C2(C1)CCCC2(C)O | 11053257 | −5.3 |
| Seychellene | CC1CCC2(C(=C)C3CCC2(C1C3)C)C | 519743 | −5.3 |
| Calarene | CC1CCC=C2C1(C3C(C3(C)C)CC2)C | 28481 | −5.3 |
| Patchouli alcohol | CC1CCC2(C(C3CCC2(C1C3)C)(C)C)O | 10955174 | −5.2 |
| Maaliol | CC1(C2C1C3C(CCCC3(C)O)(CC2)C)C | 10944069 | −5.2 |
| Desoxo-Narchinol B | CC1CCC=C2C1(C(C=CC2=O)O)C | 56835056 | −5.1 |
| Actinidine | CC1CCC2=C1C=NC=C2C | 68231 | −4.7 |
The amino acid residues involved in hydrogen bond interactions with the reference drug, ribavirin, are His421 (3.12 Å), Ser424 (2.99 Å), Gly517 (3.10 Å), Thr519 (2.98 Å) and Val538 (3.06 Å). Ribavirin also formed hydrophobic interactions with the receptor on amino acid residues Val516 and Tyr527.
The amino acid residues involved in hydrogen bond interactions with ursolic acid are His421 (3.18 Å), Val538 (2.89 Å), and Asn541 (2.96 Å). Hydrophobic interactions are formed through amino acid residues Ser424, Val515, Gly517, Thr519, and Gly523.
The amino acid residues involved in hydrogen bond interactions with nardostachys are His421 (3.03 Å), Ser424 (3.20 Å), and Gly517 (2.96 Å). Nardostachys forms hydrophobic interaction with E2 through amino acid residues Thr425, Val515, Val517, Pro525, and Tyr527.
There are no hydrogen bonds formed between α-gurjunene and E2. Hydrophobic interactions are mediated by receptor amino acid residues Val515, Val516, Gly517, Thr518, Thr519, Val538, and Trp549.
The 3-D illustration of the docking pose of phytochemicals of interest and the 2-D illustration of amino acid residues involved in the formation of hydrogen and hydrophobic interactions are represented in Figures 3-6.
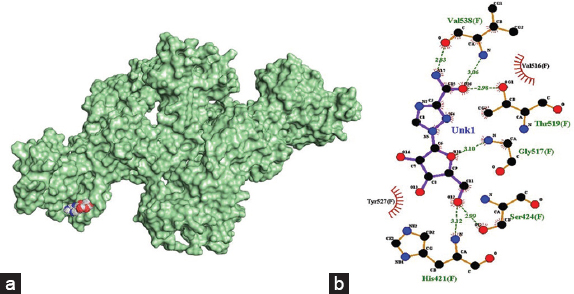 | Figure 3: (a) 3-D illustration of the docking pose of ribavirin in the binding site of viral E2 receptor, (b) 2-D illustration of amino acid residues involved in the interactions between ribavirin and E2. [Click here to view] |
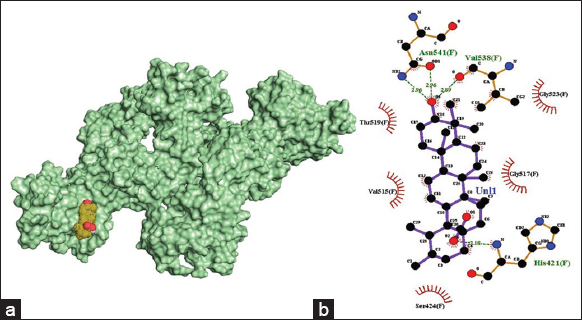 | Figure 4: (a) 3-D illustration of the docking pose of ursolic acid in the binding site of viral E2 receptor, (b) 2-D illustration of amino acid residues involved in the interactions between ursolic acid and E2. [Click here to view] |
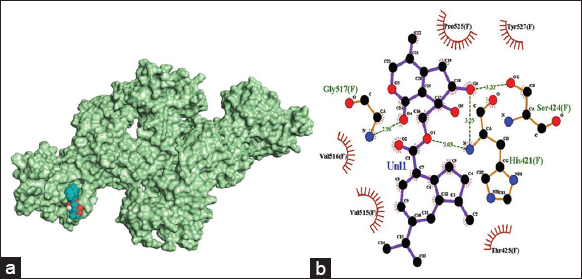 | Figure 5: (a) 3-D illustration of the docking pose of nardostachysin in the binding site of viral E2 receptor, (b) 2-D illustration of amino acid residues involved in the interactions between nardostachysin and E2. [Click here to view] |
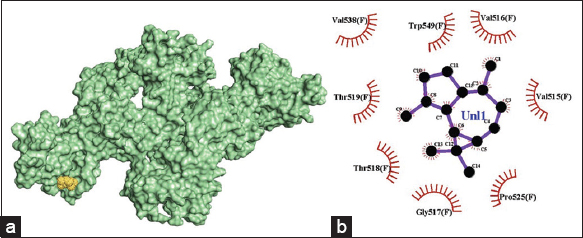 | Figure 6: (a) 3-D illustration of the docking pose of α-gurjunene in the binding site of viral E2 receptor, (b) 2-D illustration of amino acid residues involved in the interactions between α-gurjunene and E2. [Click here to view] |
The binding energies of the redocked ligands are given in supplementary material, Table S1. Docked ribavirin conformations (original docking and redocked poses) were compared using AutoDock Tools. Root mean square deviation value 0.000 Å was obtained. Hence, the credibility of docking was substantiated.
3.2. ADME Studies and BOILED-Egg Prediction Analysis
A majority of the phytochemicals showed higher binding energy than the reference drug, ribavirin. Hence, they were analyzed for their pharmacokinetic properties and drug likeliness using SwissADME. In the graphical output of the BOILED-Egg model, the points in the white ellipse (the egg white) are considered to have a high probability of passive absorption by the GI tract. The points in the yellow region (egg yolk) are highly likely to cross the BBB and access the central nervous system. Molecules not readily absorbed by the GI tract and not BBB permeant are found in the grey zone. The points colored in blue are substrates of P-glycoprotein. P-glycoprotein substrates are actively pumped out of the brain to the GI lumen. If not, they are marked as red points. The BOILED-Egg output of the phytochemicals, along with ribavirin is shown in Figure 7.
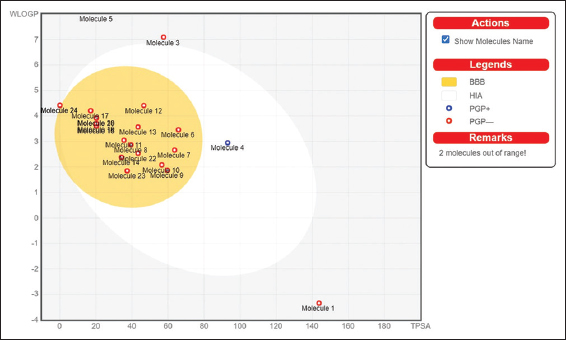 | Figure 7: BOILED-Egg output of Ribavirin and the phytochemicals of Nardostachys jatamansi (Molecule 1- Ribavirin, Molecule 2- Taraxerone, Molecule 3- Ursolic Acid, Molecule 4- Nardostachysin, Molecule 5- Lupeol, Molecule 6- Dihydrojatamansin, Molecule 7- Oroselol, Molecule 8- Seselin, Molecule 9- Jatamansinol, Molecule 10- Jatamansinone, Molecule 11- Nardosinone, Molecule 12- Jatamol B, Molecule 13- Oroselone, Molecule 14- Nardoaristolone, Molecule 15- Maaliol, Molecule 16- Seychellene, Molecule 17- Patchouli alcohol, Molecule 18- Jatamansone, Molecule 19- Spirojatamol, Molecule 20- Jatamol A, Molecule 21- Valeranol, Molecule 22- Angelicin, Molecule 23- Desoxo-Narchinol B, Molecule 24- α-Gurjunene). [Click here to view] |
The outcomes and conclusions of ADME studies of compounds are highly governed by Lipinksi’s Rule of 5 (Ro5), which defines the rules of drug-likeness. Various criteria of the Ro5 for drug candidates are given in Table 2.
Table 2: Lipinski’s rule of 5 for drugs.
| Properties | Rule |
|---|---|
| Molecular weight | <500 g/mol |
| H-bond acceptors | ≤10 |
| H-bond donors | ≤5 |
| CLOGP | <5 |
| TPSA | <140 Å |
TPSA: Total polar surface area, CLOGP: Consensus Log Po/w
For an orally active drug candidate to be considered, it must not violate more than one of the criteria of the Lipinksi’s Ro5 [32]. Abbott’s Bioavailability score is directly related to Lipinksi’s rule, such that the bioavailability score of any compound obeying the Lipinski’s rule is 0.55 [33-38].
The compound showing the highest negative binding energy against the viral E2 receptor, taraxerone, was found to be out of range of the BOILED-Egg; hence, further analysis of the specific compound was not feasible. Three compounds were selected for further analysis, namely ursolic acid, nardostachysin, and α-gurjunene. These compounds were found to be impermeant of the BBB. The reference drug, ribavirin, was shown to have a molecular weight of 244.2 g/mol with high water solubility. It has low GI absorption with one violation each for Ghose, Veber, and Egan parameters in druglikeness analysis. It also showed one violation for lead likeness as it has a molecular weight lower than 250 g/mol. In line with the detailed parameters of the study provided by SwissADME, the physicochemical properties, lipophilicity, pharmacokinetics, water solubility, druglikeness, and medicinal chemistry, the properties of the three probable candidate compounds, namely, ursolic acid, nardostachys, and α-gurjunene, were compared to Ribavirin for treatment of HCV infection. The details of ADME studies of the phytochemicals and the reference drug are given in Table 3.
Table 3: Absorption, delivery, metabolism, and excretion studies of ribavirin and the three phytochemicals considered for further studies namely ursolic acid, nardostachysin and α-gurjunene.
| Properties | Parameters | Molecules | |||
|---|---|---|---|---|---|
| Ribavirin | Ursolic acid | Nardostachysin | α-gurjunene | ||
| Physicochemical properties | Formula | C8H12N4O5 | C30H48O3 | C25H34O6 | C15H24 |
| Molecular weight (g/mol) | 244.2 | 456.7 | 430.53 | 204.35 | |
| H-bond acceptor | 7 | 3 | 6 | 0 | |
| H-bond donor | 4 | 2 | 2 | 0 | |
| TPSA (Å) | 143.72 | 57.53 | 93.06 | 0 | |
| Lipophilicity | Log Po/w (XLOGP3) | −1.85 | 7.34 | 2.78 | 4.1 |
| Log Po/w (WLOGP) | −3.34 | 7.09 | 2.95 | 4.42 | |
| Log Po/w (MLOGP) | −2.94 | 5.82 | 2.88 | 5.65 | |
| CLOGP | −2.18 | 5.88 | 3.12 | 4.49 | |
| Water solubility | Log S (ESOL) | −0.21 | −7.23 | −3.93 | −3.69 |
| ESOL class | Very soluble | Poorly soluble | Soluble | Soluble | |
| Log S (Ali) | −0.65 | −8.38 | −4.39 | −3.81 | |
| Ali Class | Very soluble | Poorly soluble | Moderately soluble | Soluble | |
| Log S (SILICOS-IT) | 1.76 | −5.67 | −3.05 | −3.52 | |
| SILICOS-IT class | Soluble | Moderately soluble | Soluble | Soluble | |
| Pharmokinetics | GI absorption | Low | Low | High | Low |
| BBB permeant | No | No | No | No | |
| Pgp substrate | No | No | Yes | No | |
| Log Kp (skin permeation) | −9.1 | −3.87 | −6.95 | −4.64 | |
| Druglikeness | Lipinski #violations | 0 | 1 | 0 | 1 |
| Ghose #violations | 1 | 3 | 0 | 0 | |
| Veber #violations | 1 | 0 | 0 | 0 | |
| Egan #violations | 1 | 1 | 0 | 0 | |
| Muegge #violations | 0 | 1 | 0 | 1 | |
| Bioavailability score | 0.55 | 0.85 | 0.55 | 0.55 | |
TPSA value ≤140 Å and H-bond acceptors and donors ≤12 are considered to have good oral bioavailability. Compounds with CLOGP >5, molecular weight >500 g/mol, H- bond acceptors >10 and H-bond >5 are considered to have poor absorption. TPSA: Total polar surface area, CLOGP: Consensus Log Po/w, BBB: Blood-brain barrier, GI: Gastrointestinal
Among the three phytochemicals selected for ADME studies, two molecules, namely nardostachysin and α-gurjunene obeyed Lipinksi’s Rule of 5 and obtained a bioavailability score of 0.55. Ursolic acid had one violation of Lipinksi’s rule as its lipophilicity is 5.88. However, it is considered for further studies as it reported the least violation of Lipinski’s rule, which can be neglected [39].
3.3. Toxicity Analysis
The toxicity of the three selected phytochemicals, namely ursolic acid, nardostachysin, and α-gurjunene, were analyzed using pkCSM, a free online web tool. The reference drug, ribavirin, was also analyzed using pkCSM, and the potential candidate compounds were compared to it. AMES toxicity assay is used to determine the mutagenicity and carcinogenicity of compounds. It is also called bacterial mutation assay, wherein the compounds are evaluated for their capacity to reverse the mutations of Salmonella typhimurium and/or Escherichia coli [40]. hERG (human ether-a-go-go related gene) and its dysfunction has been reported to induce QT prolongation, which may eventually lead to ventricular fibrillation, fainting, and cardiac death. Modern drug design techniques recognize hERG as an anti-target for drug candidates [41]. All three potential drug molecules showed no AMES toxicity, and none were found to be inhibitors of hERG I and hERG II. However, ursolic acid was assessed and confirmed to be hepatotoxic and is not a suitable candidate drug molecule. The detailed analysis of the toxicity of the three phytochemicals and ribavirin are represented in Table 4.
Table 4: Toxicity studies of ribavirin, ursolic acid, nardostachysin and α-gurjunene using pkCSM.
| Properties | Molecules | |||
|---|---|---|---|---|
| Ribavirin | Ursolic acid | Nardostachysin | α-Gurjunene | |
| AMES toxicity | No | No | No | No |
| Maximum tolerated dose (human) (log mg/kg/day) | 1.011 | 0.199 | −0.495 | 0.212 |
| hERG I inhibitor | No | No | No | No |
| hERG II inhibitor | No | No | No | No |
| Oral rat acute toxicity (LD50) (mol/kg) | 1.988 | 2.346 | 2.414 | 1.606 |
| Oral rat chronic toxicity (LOAEL) (log mg/kg_bw/day) | 3.096 | 2.054 | 62.536 | 1.371 |
| Hepatotoxicity | No | Yes | No | No |
| Skin sensitization | No | No | No | No |
| Tetrahymena pyriformis toxicity (log ug/L) | 0.285 | 0.285 | 0.331 | 1.263 |
| Minnow toxicity (mm) | 4.626 | −0.787 | 1.727 | 0.359 |
LD50 indicates the lethal dose of administered chemicals, which is expected to cause death in 50% of the experimental animals in a period [42]. The LOAEL reports oral rat chronic toxicity [43]. T. pyriformis is a unicellular, ciliated protozoan, a commonly used model for determining toxic endpoints of chemicals [44].
From Tables 1-3, we can conclude that nardostachysin and α-gurjunene are the standout phytochemicals in N. jatamansi. Having higher negative binding energies than ribavirin and satisfying all the criteria in ADME and toxicity studies, nardostachysin and α-gurjunene can be suggested as possible supplements for treating HCV infection.
4. DISCUSSION
Molecular docking aims to predict the interaction and structural complex between receptor and small molecule ligands. Molecular docking is a computer-aided drug-designing process whose outcome is based on the structure of the target receptor protein and the ligand, i.e., phytochemicals, in our study. Its efficiency, high speed, and cost-effectiveness have made molecular docking the basis of modern drug discovery. Due to these multiple characteristics, large amounts of molecules can be screened and filtered quickly, depending on the binding energies between the receptors and ligands [18,19]. Recently, it has been widely used as a part of CADD. The varying conformations of the ligands are sampled onto the active site of the protein. These conformations are then ranked through a scoring function. Ideally, conformation with the lowest or negative binding energies is chosen [18,25]. In our study, we compared the phytochemicals of N. jatamansi to an already established antiviral drug for HCV, ribavirin [2]. Should any phytochemicals have higher negative binding energy than ribavirin, they are considered for further analysis of pharmacokinetics, drug-likeness, and toxicity [26,30,31]. The process of molecular docking not only gives an accurate prediction of small molecule interactions, but is also highly efficient in terms of time consumption and cost-effectiveness. Using docking studies, piperine has also been recently reported to have potential antiviral properties against dengue and ebola viruses [45]. Piperine and curcumin could also be potential inhibitors of SARS-CoV-2 by disrupting the S-hACE2 complex [22,46,47].
Similarly, wedelolactone, a coumarin, has shown inhibiting properties against the spike protein of SARS CoV-2 [48]. Furthermore, molecular docking studies reduce the possibility of high-risk preclinical trials in the pharmaceutical industry. Secondary metabolites of Xylopia aethiopica have been reported to show promising antiviral properties against viral proteins of measles [49]. Bael fruit extract has also been reported to exhibit antitumor activity [50]. Potential inhibitors of NS2B-NS3, a viral replication protease, have also been obtained from phytochemical constituents of A. paniculata [51]. These data are obtained through molecular docking techniques.
In our study, 17 of the 25 phytochemicals found in N. jatamansi had higher negative binding energy than ribavirin and were analyzed in further studies. Ursolic acid, nardostachysin, and α-gurjunene were finalized as potential candidates for the treatment of HCV. However, following ADME and toxicity studies, nardostachysin proved to be the most promising compound, satisfying the conditions specified for clinical use as a drug.
The three phytochemicals, i.e., ursolic acid, nardostachys, and α-gurjunene, were selected because they have higher negative binding energy than ribavirin, which is widely used as an antiviral drug. In addition, from the BOILED-Egg model, the three phytochemicals were impermeant of the BBB, an essential criterion for use in clinical trials. BBB permeability has been increased by drug abuse, such as cocaine, which increases the influx of peripheral toxins into the brain [52]. Basically, compounds with high GI absorption may be supplemented through the oral route. Among the three, nardostachysin had the highest GI absorption [31]. Toxicity studies indicated that all three tested negative for inhibition of hERG I and hERG II. hERG coded protein is an important one involved in cardiac repolarization. Inhibition of hERG I and hERG II causes blockade of potassium ion channels linked to QT prolongation. Hence, hERG inhibitors block cardiac repolarization and may lead to cardiac arrhythmia [53,54]. AMES test is an assay to establish the mutagenicity and carcinogenicity of various compounds [55]. It was described by Dorothy Maron and Bruce Ames in 1983. All three compounds showed no AMES toxicity. Hepatotoxicity studies showed that ursolic acid could cause liver damage on administration. Drug-induced liver injury is one of the most frequent causes of acute liver failure. One in every 4.5 drug failures in clinical trials is attributed to hepatotoxicity of potential drug compounds [56].
N. jatamansi is an important herbal plant found in the Northern Himalayas. The extracts of the roots and rhizomes are specifically of high medicinal importance. Nardostachysin is a terpenoid ester, majorly concentrated in the rhizome of the plant, and is considered to have antitumor properties. It is also used as a supplement for hair regrowth [57]. It is also described as having nootropic activity that can improve cognitive impairment [13,58]. Although N. jatamansi is widely used as an ayurvedic medication, clinical use of the plant and its extract as a drug are still lacking [59]. However, the data from our study pointed out that nardostachysin and α-gurjunene are potential inhibitors of the viral E2 receptor and, hence, could be used as a treatment for HCV infection.
5. CONCLUSION
This study investigated the potential presence of phytochemicals in N. jatamansi to inhibit the E2 receptor of HCV. Based on the literature review, 25 phytochemicals were screened, and an antiviral drug was used to treat liver disease caused by HCV. In-silico studies used docking tools such as AutoDock Vina, PyMol, and Discovery Studio Visualizer to examine the binding affinities between phytochemicals and receptor molecules. Following this, pharmacokinetics and drug-likeness were examined using SwissADME and pkCSM. In comparison to the reference drug, ribavirin, the bioactive compounds of N. jatamansi, nardostachysin and α-gurjunene were considered to be potential inhibitors of E2 protein. They are suitable candidates as therapeutic agents that interfere with HCV life cycle. However, in vivo and in vitro studies may be carried out to validate the phytochemicals’ potential for use as inhibitors for E2 protein treatment for HCV infections.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to the conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
This study does not involve experiments on animals or human subjects.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data generated in this study are represented in table and text form within this article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Manns MP, Buti M, Gane E, Pawlotsky JM, Razavi H, Terrault N, et al. Hepatitis C virus infection. Nat Rev Dis Primers 2017;3:17006. [https://doi.org/10.1038/nrdp.2017.6]
2. Giannini C, Bréchot C. Hepatitis C virus biology. Cell Death Differ 2003;10 Suppl 1:S27-38. [https://doi.org/10.1038/sj.cdd.4401121]
3. Tong Y, Lavillette D, Li Q, Zhong J. Role of Hepatitis C virus envelope glycoprotein E1 in virus entry and assembly. Front Immunol 2018;9:1411. [https://doi.org/10.3389/fimmu.2018.01411]
4. Kong L, Giang E, Nieusma T, Kadam RU, Cogburn KE, Hua Y, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science 2013;342:1090-4. [https://doi.org/10.1126/science.1243876]
5. Who.int. Geneva:Hepatitis C;c2022. Available from:https://www.who.int/news-room/fact-sheets/detail/hepatitis-c [Last accessed on 2023 Mar 04].
6. Hepatitis.uw.edu. Seattle:Hepatitis C Online;2023. Available from:https://www.hepatitisc.uw.edu [Last accessed on 2023 Aug 03].
7. Cowton VM, Angus AG, Cole SJ, Markopoulou CK, Owsianka A, Dunlop JI, et al. Role of conserved E2 residue W420 in receptor binding and hepatitis C virus infection. J Virol 2016;90:7456-68. [https://doi.org/10.1128/JVI.00685-16]
8. Ashfaq UA, Javed T, Rehman S, Nawaz Z, Riazuddin S. An overview of HCV molecular biology, replication and immune responses. Virol J 2011;8:161. [https://doi.org/10.1186/1743-422X-8-161]
9. Morozov VA, Lagaye S. Hepatitis C virus:Morphogenesis, infection and therapy. World J Hepatol 2018;10:186-212. [https://doi.org/10.4254/wjh.v10.i2.186]
10. Berggren KA, Suzuki S, Ploss A. Animal models used in hepatitis C virus research. Int J Mol Sci 2020;21:3869. [https://doi.org/10.3390/ijms21113869]
11. Chiou HE, Liu CL, Buttrey MJ, Kuo HP, Liu HW, Kuo HT, et al. Adverse effects of ribavirin and outcome in severe acute respiratory syndrome:Experience in two medical centers. Chest 2005;128:263-72. [https://doi.org/10.1378/chest.128.1.263]
12. Kaur H, Lekhak MM, Chahal S, Goutam U, Jha P, Naidoo D. Nardostachys jatamansi (D.Don) DC.:An invaluable and constantly dwindling resource of the Himalayas. S Afr J Bot 2020;135:252-67. [https://doi.org/10.1016/j.sajb.2020.08.010]
13. Karkada G, Shenoy KB, Halahalli H, Karanth KS. Nardostachys jatamansi extract prevents chronic restraint stress-induced learning and memory deficits in a radial arm maze task. J Nat Sci Biol Med 2012;3:125-32. [https://doi.org/10.4103/0976-9668.101879]
14. Khan AM, Singh VK, Agnihotri NK, Joshi MC, Kumar K. Secondary metabolites of genus nardostachys and their medicinal importance- recent updates. Am J Phytomed Clin Ther 2021;9:10.
15. Dhiman N, Bhattacharya A. Nardostachys jatamansi (D.Don) DC.-Challenges and opportunities of harnessing the untapped medicinal plant from the Himalayas. J Ethnopharmacol 2020;246:112211. [https://doi.org/10.1016/j.jep.2019.112211]
16. Chauhan HK, Oli S, Bisht AK, Meredith C, Leaman D. Review of the biology, uses and conservation of the critically endangered endemic Himalayan species Nardostachys jatamansi (Caprifoliaceae). Biodivers Conserv 2021;30:3315-33. [https://doi.org/10.1007/s10531-021-02269-6]
17. Pant HC, Pant HV, Kumar A, Tomar H, Dev Sharma M, Gaurav N. In vitro clonal propagation of Nardostachys jatamansi:A traditional himalayan medicinal plant. J Mountain Res 2021;16:87-98. [https://doi.org/10.51220/jmr.v16i3.10]
18. Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking:A powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 2011;7:146-57. [https://doi.org/10.2174/157340911795677602]
19. Trott O, Olson AJ. AutoDock vina:Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61. [https://doi.org/10.1002/jcc.21334]
20. Basu A, Sarkar A, Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci Rep 2020;10:17699. [https://doi.org/10.1038/s41598-020-74715-4]
21. Nag A, Banerjee R, Chowdhury RR, Krishnapura Venkatesh C. Phytochemicals as potential drug candidates for targeting SARS CoV 2 proteins, an in silico study. Virusdisease 2021;32:98-107. [https://doi.org/10.1007/s13337-021-00654-x]
22. Nag A, Paul S, Banerjee R, Kundu R. In silico study of some selective phytochemicals against a hypothetical SARS-CoV-2 spike RBD using molecular docking tools. Comput Biol Med 2021;137:104818. [https://doi.org/10.1016/j.compbiomed.2021.104818]
23. Rajpoot S, Solanki K, Kumar A, Zhang KY, Pullamsetti SS, Savai R, et al. In-silico design of a novel tridecapeptide targeting spike protein of SARS-CoV-2 variants of concern. Int J Pept Res Ther 2022;28:28. [https://doi.org/10.1007/s10989-021-10339-0]
24. Qawoogha SS, Shahiwala A. Identification of potential anticancer phytochemicals against colorectal cancer by structure-based docking studies. J Recept Signal Transduct Res 2020;40:67-76. [https://doi.org/10.1080/10799893.2020.1715431]
25. Hariprasath R, Akashpriya C, Lakshmaiah VV, Praveen N. In silico studies of viral protein inhibitors of Marburg virus using phytochemicals from Andrographis paniculata. J Appl Biol Biotechnol 2022;11:151-60. [https://doi.org/10.7324/JABB.2023.110121]
26. Diniz WJ, Canduri F. Review-article bioinformatics:An overview and its applications. Genet Mol Res 2017;16:1-21. [https://doi.org/10.4238/gmr16019645]
27. Daina A, Michielin O, Zoete V. SwissADME:A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7:42717. [https://doi.org/10.1038/srep42717]
28. Pires DE, Blundell TL, Ascher DB. pkCSM:Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 2015;58:4066-72. [https://doi.org/10.1021/acs.jmedchem.5b00104]
29. Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock vina 1.2.0:New docking methods, expanded force field, and python bindings. J Chem Inf Model 2021;61:3891-8. [https://doi.org/10.1021/acs.jcim.1c00203]
30. Cheng F, Li W, Liu G, Tang Y. In silico ADMET prediction:Recent advances, current challenges and future trends. Curr Top Med Chem 2013;13:1273-89. [https://doi.org/10.2174/15680266113139990033]
31. Daina A, Zoete V. A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016;11:1117-21. [https://doi.org/10.1002/cmdc.201600182]
32. Benet LZ, Hosey CM, Ursu O, Oprea TI. BDDCS, the rule of 5 and drugability. Adv Drug Deliv Rev 2016;101:89-98. [https://doi.org/10.1016/j.addr.2016.05.007]
33. Martin YC. A bioavailability score. J Med Chem 2005;48:3164-70. [https://doi.org/10.1021/jm0492002]
34. Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 2002;45:2615-23. [https://doi.org/10.1021/jm020017n]
35. Amin ML. P-glycoprotein inhibition for optimal drug delivery. Drug Target Insights 2013;7:27-34. [https://doi.org/10.4137/DTI.S12519]
36. Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2012;64:4. [https://doi.org/10.1016/j.addr.2012.09.019]
37. Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem 1999;1:55-68. [https://doi.org/10.1021/cc9800071]
38. Zafar F, Gupta A, Thangavel K, Khatana K, Sani AA, Ghosal A, et al. Physicochemical and pharmacokinetic analysis of anacardic acid derivatives. ACS Omega 2020;5:6021-30. [https://doi.org/10.1021/acsomega.9b04398]
39. Mahgoub RE, Atatreh N, Ghattas MA. Chapter three - using filters in virtual screening:A comprehensive guide to minimize errors and maximize efficiency. Annual Rep Med Chem 2022;59:99-136. [https://doi.org/10.1016/bs.armc.2022.09.002]
40. Pan X. Mutagenicity evaluation of nanoparticles by the ames assay. Methods Mol Biol 2021;2326:275-85. [https://doi.org/10.1007/978-1-0716-1514-0_20]
41. Creanza TM, Delre P, Ancona N, Lentini G, Saviano M, Mangiatordi GF. Structure-based prediction of hERG-related cardiotoxicity:A benchmark study. J Chem Inf Model 2021;61:4758-70. [https://doi.org/10.1021/acs.jcim.1c00744]
42. Minerali E, Foil DH, Zorn KM, Ekins S. Evaluation of assay central machine learning models for rat acute oral toxicity prediction. ACS Sustain Chem Eng 2020;8:16020-7. [https://doi.org/10.1021/acssuschemeng.0c06348]
43. Gadaleta D, Marzo M, Toropov A, Toropova A, Lavado GJ, Escher SE, et al. Integrated in silico models for the prediction of no-observed-(Adverse)-effect levels and lowest-observed-(adverse)-effect levels in rats for sub-chronic repeated-dose toxicity. Chem Res Toxicol 2021;34:247-57. [https://doi.org/10.1021/acs.chemrestox.0c00176]
44. Yoshioka Y, Ose Y, Sato T. Testing for the toxicity of chemicals with Tetrahymena pyriformis. Sci Total Environ 1985;43:149-57. [https://doi.org/10.1016/0048-9697(85)90037-3]
45. Nag A, Chowdhury RR. Piperine, an alkaloid of black pepper seeds can effectively inhibit the antiviral enzymes of Dengue and Ebola viruses, an in silico molecular docking study. Virusdisease 2020;31:308-15. [https://doi.org/10.1007/s13337-020-00619-6]
46. Nag A, Banerjee R, Paul S, Kundu R. Curcumin inhibits spike protein of new SARS-CoV-2 variant of concern (VOC) Omicron, an in silico study. Comput Biol Med 2022;146:105552. [https://doi.org/10.1016/j.compbiomed.2022.105552]
47. Rattis BA, Ramos SG, Celes MR. Curcumin as a potential treatment for COVID-19. Front Pharmacol 2021;12:675287. [https://doi.org/10.3389/fphar.2021.675287]
48. Katuwal S, Upadhyaya SR, Marahatha R, Shrestha A, Regmi BP, Khadayat K, et al. In silico study of coumarins:Wedelolactone as a potential inhibitor of the spike protein of the SARS-CoV-2 variants. J Trop Med 2023;2023:47717445. [https://doi.org/10.1155/2023/4771745]
49. Oloche JJ, Oluremi BB, Aruwa CE, Sabiu S. Molecular modeling identification of key secondary metabolites from Xylopia aethiopica as promising therapeutics targeting essential measles viral proteins. Evid Based Complement Alternat Med 2023;2023:1575358. [https://doi.org/10.1155/2023/1575358]
50. Hemakumar C, Ravindranath BS, Ravishankar GA, Ramirez DC, Kiran SV. Marmesin and marmelosin interact with the heparan sulfatase-2 active site:Potential mechanism for phytochemicals from bael fruit extract as antitumor therapeutics. Oxid Med Cell Longev 2023;2023:9982194. [https://doi.org/10.1155/2023/9982194]
51. Thirumoorthy G, Tarachand SP, Nagella P, Veerappa Lakshmaiah V. Identification of potential ZIKV NS2B-NS3 protease inhibitors from Andrographis paniculata:An insilico approach. J Biomol Struct Dyn 2021;40:11203-15. [https://doi.org/10.1080/07391102.2021.1956592]
52. Pimentel E, Sivalingam K, Doke M, Samikkannu T. Effects of drugs of abuse on the blood-brain barrier:A brief overview. Front Neurosci 2020;14:513. [https://doi.org/10.3389/fnins.2020.00513]
53. Danielsson BR, Lansdell K, Patmore L, Tomson T. Phenytoin and phenobarbital inhibit human HERG potassium channels. Epilepsy Res 2003;55:147-57. [https://doi.org/10.1016/S0920-1211(03)00119-0]
54. Danker T, Möller C. Early identification of hERG liability in drug discovery programs by automated patch clamp. Front Pharmacol 2014;5:203. [https://doi.org/10.3389/fphar.2014.00203]
55. Maron DM, Ames BN. Revised methods for the Salmonella mutagenicity test. Mutat Res 1983;113:173-215. [https://doi.org/10.1016/0165-1161(83)90010-9]
56. Dirven H, Vist GE, Bandhakavi S, Mehta J, Fitch SE, Pound P, et al. Performance of preclinical models in predicting drug-induced liver injury in humans:A systematic review. Sci Rep 2021;11:6403. [https://doi.org/10.1038/s41598-021-85708-2]
57. Chatterjee A, Basak B, Saha M, Dutta U, Mukhopadhyay C, Banerji J, et al. Structure and stereochemistry of nardostachysin, a new terpenoid ester constituent of the rhizomes of Nardostachys jatamansi. J Nat Prod 2000;63:1531-3. [https://doi.org/10.1021/np990503m]
58. Kulkarni R, Girish KJ, Kumar A. Nootropic herbs (Medhya Rasayana) in Ayurveda:An update. Pharmacogn Rev 2012;6:147-53. [https://doi.org/10.4103/0973-7847.99949]
59. Drugs.com. Auckland:Jatamansi;c2022. Available from:https://www.drugs.com/npp/jatamansi.html [Last accessed on 2023 Mar 02].