1. INTRODUCTION
MicroRNAs (miRNAs) are a type of non-coding RNAs with about 22 nucleotides that interact with mRNAs to regulate gene expression post-transcriptionally [1]. Over the past decade, non-coding RNAs have been discovered to have a regulatory role on cellular molecules such as adaptor molecules, signaling molecules, and receptor protein in both cellular and metabolic signalling [2]. Small non-coding molecules (21–23-nucleotide) called miRNAs have been found to be present in the circulatory system. MiRNAs have been investigated and identified to be complementary to messenger RNA (mRNA) and they play an important role in cellular and molecular signaling either by repression or degradation of mRNA, which codes for specific molecules [3]. These can post-transcriptionally regulate the gene expression and can also regulate different cellular processes including insulin synthesis and secretion, pancreatic β cell development and function, insulin signaling, and glucose homeostasis.
Diabetes mellitus is a progressive metabolic disorder that is distinguished by elevated blood sugar levels and poses a serious threat to human health. Understanding the underlying processes involved in the initiation and development of Type 2 diabetes mellitus (T2DM) is a better strategy in preventing T2DM-related mortality [4]. Pre-diabetes is an intermediate stage of diabetes that occurs between normal glucose tolerance and T2DM and is caused by dysregulated functioning of β cells and increased insulin resistance. It is determined by the occurring impaired fasting glucose and/or impaired glucose tolerance [5]. Dysregulation of miRNA can affect the functioning of different tissues during the progression of T2DM [6]. Our study aims to identify these dysregulated circulatory miRNAs that can serve as potential biomarkers for the progression of pre-diabetes to T2DM and to analyze the candidate miRNA-target interactions in the insulin signal transduction pathway at the molecular level by using in silico approach.
2. MATERIALS AND METHODS
2.1. Identifying Target Genes and miRNAs
First, from the available literature, insulin receptor (INSR) gene and the INSR substrate 1 (IRS-1) gene were selected as the targets. Further, online databases including miRDB (http://mirdb.org/), TargetScanHuman ver.7.1 (http://www.targetscan.org), DIANA-microT-CDS web server (http://diana.imis.athena-innovation.gr), and miRmap (https://mirmap.ezlab.org/) were thoroughly evaluated for selecting miRNAs having target genes INSR and IRS-1. The selection was based on the target prediction scores which suggest the confidence level for target prediction. The threshold valve for the target prediction was set to >70%. The greater the target prediction score, the more confidence in the prediction.
2.2. miRNA-Target Gene Functional Enrichment Annotation and Interaction Network
MIENTURNET (http://userver.bio.uniroma1.it/apps/mienturnet/) is the web server used to execute miRNA-target gene enrichment analysis and build their interactions network. MIENTURNET searches for mRNAs targeted by input list of candidate miRNAs, and it performs a statistical evaluation for over-representation of miRNA-target interactions together with a network-based visualization and analysis of the resulting enriched interactions [7]. The enrichment analysis was performed by setting a threshold for the minimum number of miRNA-target interactions as default value of 2 and the adjusted P-values as 1. By clicking on miRTarBase tab, the experimentally confirmed miRNA-target interactions were considered. Functional enrichment analysis of the respective target genes of selected miRNAs was executed using KEGG annotation database. The KEGG represents a database consisting of known functional genes along with their respective biological and biochemical pathways.
2.3. Prediction of miRNA-mRNA Duplex Structure
To corroborate the binding affinity between the miRNAs and their corresponding target genes, secondary structures were predicted and the minimum free energy (MFE) scores of duplex interactions were computed using RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The conserved sites with higher PCT values obtained from TargetScan Human ver.7.1 were selected to construct the duplex structures. The formation of the duplex structure suggested that the duplexes formed are energetically favorable and the genes are functional. The results obtained for the miRNA-mRNA duplex were displayed in a dot-bracketed format.
2.4. miRNA-mRNA Molecular Docking Studies
The mRNA of the target gene complementary to the selected miRNAs was docked using the online docking server HNADOCK (http://huanglab.phys.hust.edu.cn/hnadock/). HNADOCK server makes use of HDOCKlite, an FFT-based global docking program that samples the supposed binding of one nucleic acid to another and uses optimized shape-linked pairwise scores for grid-associated matching [8]. Finally, the docked complexes were visualized using BIOVIA Discovery Studio visualizer 2019.
3. RESULTS
3.1. Selection and Screening of miRNAs
From the literature survey conducted, 17 miRNAs were found to be targeting the genes INSR and IRS-1 and were expressed during the development of T2DM. Based on target prediction scores obtained from the databases miRDB, TargetScan, DIANA-microT-CDS, and miRmap showed 7 miRNAs − i.e., miR-15b-5p, miR-195-5p, miR-27b-3p, miR-128-3p, miR-379-5p, miR-424-5p, miR-29b-1-5p targeting INSR and 11 miRNAs miR-15b-5p, miR-195-5p, miR-27b-3p, miR-128-3p, miR-30d-5p, miR-143-5p, miR-33b-5p, miR-96-5p, miR-144-3p, miR-424-5p, miR-7-5p, miR-589-5p, and miR-30d-5p targeting IRS-1 that had a target prediction score >70% [Supplementary Table 1].
3.2. Functional Annotation and Network Analysis
miRNA-target network analysis showed miR-15b-5p and miR-195-5p had closer interaction towards target gene INSR and miR-144-3p, miR-148- 3p, miR-7-5p had closer interaction with target gene IRS-1 [Figure 1]. Functional enrichment analysis was performed with target genes of miR-15b-5p, miR-195-5p, miR-7-5p, miR-144-3p, and miR-148a-3p using KEGG database result (P < 0.05 is considered as statistically significant) [Table 1]. The dot plot obtained from KEGG pathway showed the selected miRNAs targeted the genes found in the PI3K-Akt pathway [Figure 2].
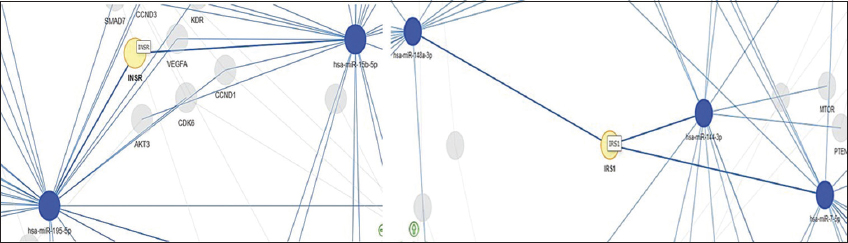 | Figure 1: Visualization of miRNA-target interaction to identify those genes that are targeted by multiple miRNAs under investigation. miRNAs are represented by blue nodes, while their target genes are represented by yellow nodes – (Based on the strong evidence category for considering strong experimental methods used by miRTarBase for validating miRNA-target interactions). [Click here to view] |
Table 1: The genes targeted by miRNAs in PI3K-Akt signaling pathway.
| miRNA | FDR | Genes |
|---|---|---|
| miR-15b-5p | 0.00000495 | CCNE1/BCL2/CCND1/VEGFA/KDR/AKT3/INSR/CCND3 |
| miR-195-5p | 1.10e-9 | CDK6/CCND1/VEGFA/BCL2/RAF1/CCND3/CDK4/CCNE1/MYB/RPS6KB1/FGF2/INSR/KDR |
| miR-7-5p | 1.47e-7 | EGFR/IRS1/RAF1/IGF1R/PIK3CD/PTK2/BCL2/RELA/CCNE1 |
| miR-144-3p | 0.00714 | MTOR/PTEN/MET/IRS1 |
| miR-148a-3p | 0.0109 | IRS1/BCL2/MET/BCL2L11 |
 | Figure 2: Dot plot of functional enrichment annotation for target genes of selected miRNAs. The color of the dots represents the adjusted P-values (FDR) (i.e., red dot represents adj P < 0.05), whereas the size of the dots represents gene ratio (i.e., the number of enriched miRNA targets each category over the number of total genes related to that category). [Click here to view] |
3.3. Prediction of Duplex between miRNAs and mRNA of INSR and IRS-1 Genes
The sequences for miR-mRNA duplex were extracted from TargetScanHuman. From the duplex sequences, the secondary folding pattern and dot-bracket structures were obtained from RNAfold web server. Based on secondary structure analysis, the MFE values were obtained [Supplementary Table 2]. The predicted MFE score indicated that target genes INSR and IRS-1 have a strong affinity towards the selected miRNAs [Table 2]. Hence, these genes and miRNAs were taken into consideration to investigate their molecular interactions.
Table 2: Secondary folding pattern of miRNA-mRNA duplexes with their predicted MFE scores using RNAfold web server.
| miRNAs | Target gene | Dot bracket | Secondary structure of miRNA-mRNA duplex | MFE score (kcal/moL) |
|---|---|---|---|---|
| miR-15b-5p | INSR | UAGCAGCACAUCAUGGUUUACAAAAUUGACCAAUAGCUGCUGCUU .(((((((.(((((.))))).))))))). |  | −4.70 |
| miR-195-5p | UAGCAGCACAGAAAUAUUGGCAAAUUGACCAAUAGCUGCUGCUU .(((((((.((((((((.)).)))))).))))))). |  | −14.60 | |
| miR-30d-5p | IRS1 | UGUAAACAUCCCCGACUGGAAGGCAUCCAUUUCAGUUUGUUUACU .(((((((.((((((((((.)).))))))))))))))). |  | −13.40 |
| miR-424-5p | CAGCAGCAAUUCAUGUUUUGAAAAGAUACAUUUCAUCUGCUGCUG ((((((((.((((.((.)).)))).)))))))) | 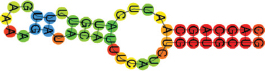 | −12.60 |
3.4. Interaction between miRNAs and mRNAs of INSR and IRS-1 Gene at the Molecular Level
After the predicted miRNA-mRNA secondary complex, the miRNAs were docked with the target genes INSR and IRS-1 using HNADOCK web server. The docking scores and ligand RMSD (Å) obtained were used for approximately predicting the binding free energy and binding affinity between the docked complexes [Table 3]. The best docked complexes with lowest docked scores and ligand RMSD that suggested a strong molecular interaction between them were downloaded from the HNADOCK and their 3D structure were visualized using BIOVIA Discovery Studio visualizer 2019 [Figure 3]. miR-15b-5p, miR-195-5p, and miR-424-5p interacting with target gene INSR had the least docking score of −340.57 kcal/moL, −310.62 kcal/moL, and −279.77 kcal/moL. Similarly, miR-424-5p, miR-15b-5p, miR-195-5p, and miR-30d-5p interacted with target gene IRS-1 with the docking score −320.94 kcal/moL, −315.12 kcal/moL, −315.12 kcal/moL, and −252.05 kcal/moL, respectively.
Table 3: Docking scores between miRNA and target mRNA using HNADOCK.
| miRNA | Target | Docking score (kcal/moL) | Ligand RMSD (Å) |
|---|---|---|---|
| Ref_let-7a-5p | INSR | −237.06 | 34.77 |
| miR-15b-5p | INSR | −340.57 | 33.61 |
| miR-195-5p | −310.62 | 49.80 | |
| miR-424-5p | −279.77 | 18.14 | |
| miR-27b-3p | −289.61 | 338.37 | |
| miR-128-3p | −222.17 | 341.94 | |
| miR-379-5p | −197.55 | 172.32 | |
| miR-424-5p | IRS-1 | −320.94 | 42.32 |
| miR-15b-5p | −315.12 | 41.62 | |
| miR-195-5p | −315.12 | 41.62 | |
| miR-30d-5p | −252.05 | 86.13 | |
| miR-144-3p | −202.23 | 117.45 | |
| miR-143-5p | −204.80 | 119.05 | |
| miR-7-5p | −260.96 | 152.20 | |
| miR-27b-3p | −262.73 | 343.39 | |
| miR-128-3p | −262.7 | 343.39 | |
| miR-96-5p | −191.78 | 415.72 |
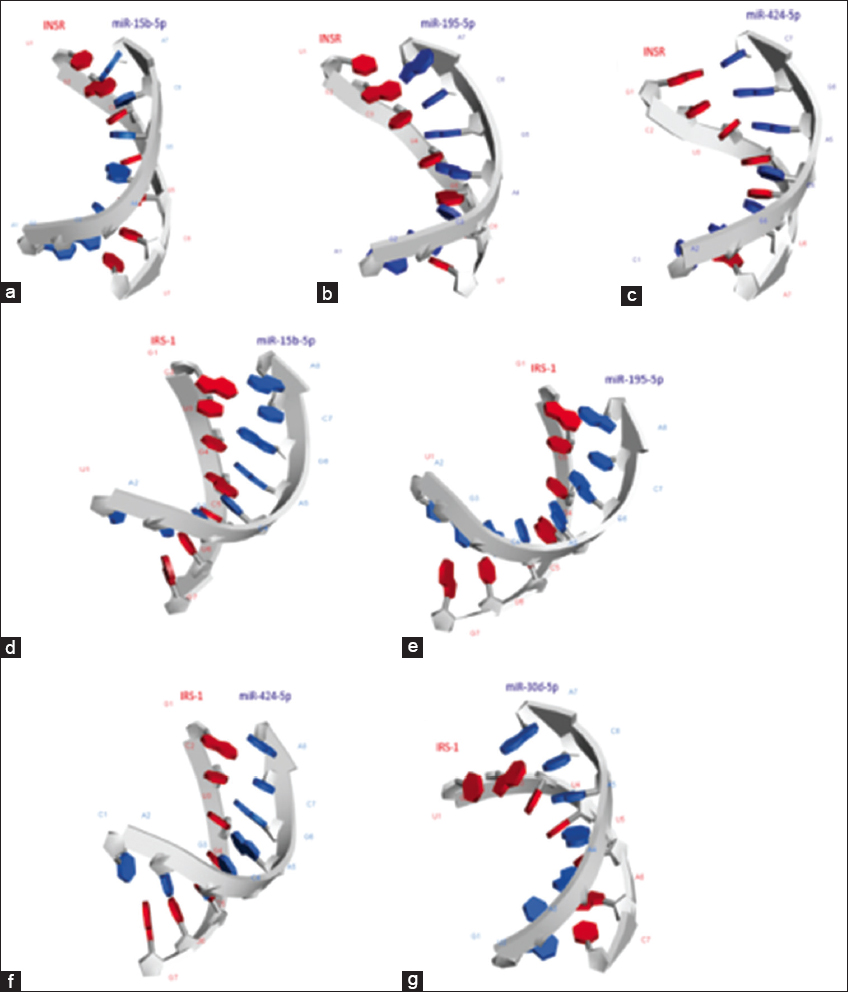 | Figure 3: Three-dimensional molecular interaction modes of (a) miR-15b-5p and mRNA of INSR gene, (b) miR-195-5p and mRNA of INSR gene, (c) miR-424-5p and mRNA of INSR gene, (d) miR-15b-5p and mRNA of IRS1 gene, (e) miR-195-5p and mRNA of IRS1 gene, and (f) miR-424-5p and mRNA of IRS1 gene, and (g) miR-30d-5p and mRNA of IRS1 gene. [Click here to view] |
4. DISCUSSION
The current research sheds light on in-silico validation and recognition of candidate miRNAs and their target genes in pre-diabetes and T2DM. Adult-onset insulin resistance causes T2DM, which is characterized by inadequate insulin action. miRNAs are known to regulate various metabolic processes such as insulin production and its release from pancreatic β islets, insulin action, and glucose homeostasis. Several miRNAs are expressed in the insulin signaling pathway and regulate the INSRs, insulin-like growth factors (IGFs), its receptors (IGFRs), and the significant protein cascades that affect insulin resistance. The loss of INSR tissue content in diabetes is one of the major contributors of impaired insulin signaling and diabetes progression [9]. IRS-1, which belongs to the IRS protein substrate family, is an endogenous substrate of the INSR expressed in insulin sensitive tissues and is also considered to be involved in the insulin-signaling pathway. IRS-1 gene variants have been linked to insulin resistance, obesity, and T2DM [10]. Therefore, INSR and IRS-1 were selected as the target genes for the study.
17 miRNAs which were found to be dysregulated in diabetic condition from the literature survey were selected for screening by target prediction using online databases. Out of the 17 miRNAs, 7 miRNAs - miR-15b-5p, miR-195-5p, miR-27b-3p, miR-128-3p, miR-379-5p, miR-424-5p, and miR-29b-1-5p were found to have target sites in INSR gene and 11 miRNAs -miR-15b-5p, miR-195-5p, miR-27b-3p, miR-128-3p, miR-30d-5p, miR-143-5p, miR-33b-5p, miR-96-5p, miR-144-3p, miR-424-5p, miR-7-5p, miR-589-5p, and miR-30d-5p had target sites in IRS-1 gene with target prediction score greater than 70 [Supplementary Table 1].
The results from the functional enrichment analysis suggested that miR-15b-5p, miR-195-5p, miR-144-3p, miR-148a-3p, and miR-7-5p (with adjusted P-value (FDR) <0.05) targeted multiple genes including INSR and IRS-1 present in the PI3K-Akt signaling pathway. miR-target network analysis showed that miR-15b-5p and miR-195-5p had closer interaction toward target gene INSR and miR-144-3p, miR-148-3p, and miR-7-5p had closer interaction with target gene IRS-1.
The validation of binding affinity between the miRNAs and their target genes was obtained from the MFE scores of miRNA-mRNA duplex structures. The duplex structure is formed when there is a stronger miRNA-mRNA binding due to the lower free energy of the complementary sequences [11]. The more negative the MFE of the duplex structures, the greater the binding affinity [12]. The optimum free energy of miRNA-target gene duplex was considered to be lower than −17 kcal/moL [13]. The resulting MFE scores of miR-15b-5p-INSR mRNA and miR-195-5p-INSR mRNA duplex structures were −14.70 kcal/moL and −14.60 kcal/moL, respectively; Similarly, MFE scores of miR-424-5p-IRS-1 mRNA and miR-30d-5p-IRS-1 mRNA were −12.60 kcal/moL and −13.40 kcal/moL suggested a stronger binding affinity between these miRNAs and their targets [Table 2].
Furthermore, to study the miRNA-target gene interaction at the molecular level, the molecular docking study was performed. The docking scores and ligand RMSD obtained were used for approximately predicting the binding free energy and binding affinity between the docked complexes. The docked complexes with lowest docked scores, namely – miR-15b-5p, miR-195-5p, and miR-424-5p having target gene INSR had the docking score of −340.57 kcal/moL, −310.62 kcal/moL, and −279.77 kcal/moL; similarly, miR-424-5p, miR-15b-5p, miR-195-5p, and miR-30d-5p interacted with target gene IRS-1 with the docking score −320.94 kcal/moL, −315.12 kcal/moL, −315.12 kcal/moL, and −252.05 kcal/moL, respectively, suggested a strong binding affinity and can be considered for further clinical analysis. Based on in-silico analysis, it is evident that INSR is the target gene for miR-15b-5p/miR-195-5p/miR-424-5p and IRS-1 is the target gene for miR-15b-5p/miR-195-5p/miR-424-5p and miR-30d-5p.
5. CONCLUSION
In this study, we could categorize miRNAs that could target the INSR and IRS-1 located upstream of the insulin signal transduction pathway, besides could be used to explore their clinical significance in pre-diabetes and T2DM. Consequently, miRNA expression studies need to be carried out to understand the clinical significance of these candidate miRNAs as possible biomarkers for early detection of pre-diabetes to facilitate new therapeutic strategies for the treatment of pre-diabetes and T2DM.
6. AUTHORS’ CONTRIBUTIONS
Sujatha Sundaresan: Conceived the concept, designed the work and analyzed the data, manuscript correction. Angel Mendonca: Executed the work and collected the data and manuscript writing. Prabu Thandapani: Took part in data collection and result discussion. Priya Swaminathan: Analyses of data, software selection. All authors have read and approved the final manuscript.
7. CONFLICTS OF INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
8. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
9. DATA AVAILABILITY
The software used in this study are mentioned in materials and methods section. The data, miRNA, and target sequences are obtained from TargetScanHuman ver.7.1.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. ACKNOWLEDGMENTS
The Article Processing Charge (APC) is supported by the National Conference on “Current Advances in Life Sciences” – Virtual Mode (NCCALS – 2022), SRM Institute of Science and Technology, Kattankulathur, Tamil Nadu, India.
REFERENCES
1. Felekkis K, Touvana E, Stefanou C, Deltas C. MicroRNAs:A newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010;14:236-40.
2. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell 2011;43:904-14. [CrossRef]
3. Orang AV, Safaralizadeh R, Kazemzadeh-Bavili M. Mechanisms of miRNA-mediated gene regulation from common downregulation to mRNA-specific upregulation. Int J Genomics 2014;2014:970607. [CrossRef]
4. Nyenwe EA, Jerkins TW, Umpierrez GE, Kitabchi AE. Management of Type 2 diabetes:Evolving strategies for the treatment of patients with Type 2 diabetes. Metabolism 2011;60:1-23. [CrossRef]
5. Tabák AG, Herder C, Rathmann W, Brunner EJ, Kivimäki M. Prediabetes:A high-risk state for diabetes development. Lancet 2012;379:2279-90. [CrossRef]
6. He Y, Ding Y, Liang B, Lin J, Kim TK, Yu H, et al. A systematic study of dysregulated MicroRNA in Type 2 diabetes mellitus. Int J Mol Sci 2017;18:456. [CrossRef]
7. Licursi V, Conte F, Fiscon G, Paci P. MIENTURNET:An interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinformatics 2019;20:545. [CrossRef]
8. He J, Wang J, Tao H, Xiao Y, Huang SY. HNADOCK:A nucleic acid docking server for modeling RNA/DNA-RNA/DNA 3D complex structures. Nucleic Acids Res 2019;47:W35-42. [CrossRef]
9. Wang Y, Zhou H, Palyha O, Mu J. Restoration of insulin receptor improves diabetic phenotype in T2DM mice. JCI Insight 2019;4:e124945. [CrossRef]
10. Alharbi KK, Khan IA, Munshi A, Alharbi FK, Al-Sheikh Y, Alnbaheen MS. Association of the genetic variants of insulin receptor substrate 1 (IRS-1) with Type 2 diabetes mellitus in a Saudi population. Endocrine 2014;47:472-7. [CrossRef]
11. Huang Y, Zou Q, Song H, Song F, Wang L, Zhang G, et al. A study of miRNAs targets prediction and experimental validation. Protein Cell 2010;1:979-86. [CrossRef]
12. Rath SN, Das D, Konkimalla VB, Pradhan SK. In silico study of miRNA based gene regulation, involved in solid cancer, by the assistance of argonaute protein. Genomics Inform 2016;14:112-24. [CrossRef]
13. Takane K, Fujishima K, Watanabe Y, Sato A, Saito N, Tomita M, et al. Computational prediction and experimental validation of evolutionarily conserved microRNA target genes in bilaterian animals. BMC Genomics 2010;11:101. [CrossRef]
APPENDIX A. SUPPLEMENTARY DATA
Supplementary Table 1: miRNA-Target Prediction scores obtained from miRNA databases.
| Target prediction score | ||||
|---|---|---|---|---|
| miRNA | miRDB | TargetScan | microT-CDS | miRmap |
| hsa-miR-15b-5p | 94 | 96 | 95 | 99.82 |
| hsa-miR-195-5p | 91 | 96 | 94.4 | 99.74 |
| hsa-miR-27b-3p | 86 | 71 | 90 | - |
| hsa-miR-128-3p | 75 | - | 75 | - |
| hsa-miR-379-5p | 73 | - | 82 | 90 |
| hsa-miR-34a-3p | - | - | - | - |
| hsa-miR-30d-5p | - | - | - | - |
| hsa-miR-143-5p | - | - | - | - |
| hsa-miR-33b-5p | - | - | - | - |
| hsa-miR-96-5p | - | - | - | - |
| hsa-miR-144-3p | - | - | - | - |
| hsa-miR-424-5p | 90 | 96 | 97 | 99.79 |
| hsa-miR-7-5p | - | - | - | 95 |
| hsa-miR-29b-1-5p | 77 | - | 88 | 98 |
| hsa-miR-589-5p | - | - | - | - |
| hsa-miR-148a-3p | - | - | - | - |
| hsa-miR-30a-5p | - | - | - | - |
| Target gene: Insulin receptor substrate (IRS-1) | ||||
| hsa-miR-15b-5p | 71 | 78 | 77 | 90 |
| hsa-miR-195-5p | 75 | 78 | 79 | 91 |
| hsa-miR-27b-3p | 73 | 78 | 98 | 72 |
| hsa-miR-128-3p | 96 | 91 | 100 | - |
| hsa-miR-379-5p | - | - | - | 72 |
| hsa-miR-34a-3p | - | - | - | - |
| hsa-miR-30d-5p | 87 | - | 91 | - |
| hsa-miR-143-5p | 78 | - | 81 | 90 |
| hsa-miR-33b-5p | 73 | - | 79 | 81 |
| hsa-miR-96-5p | 89 | 85 | 93 | 90 |
| hsa-miR-144-3p | 78 | 87 | 74 | 75 |
| hsa-miR-424-5p | - | 78 | 91 | 92 |
| hsa-miR-7-5p | 95 | 86 | 100 | 96 |
| hsa-miR-29b-1-5p | - | - | - | - |
| hsa-miR-589-5p | - | - | 88 | 90 |
| hsa-miR-148a-3p | - | - | - | 89 |
| hsa-miR-30a-5p | 87 | 71 | 90 | 84 |
Supplementary Table 2: Predicted secondary structures of miRNA-mRNA duplex along with the MFE score.
| miRNAs | Target gene | Dot bracket | Secondary structure of miRNA-mRNA duplex | MFE score (kcal/mol) |
|---|---|---|---|---|
| Ref_let-7a-5p | INSR | UGAGGUAGUAGGUUGUAUAGUUUU UUUUCAGCACAGUCUACCUCA ((((((((.((((.)))).)))))))) | 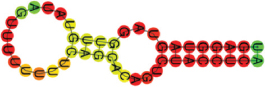 | −12.90 |
| miR-15b-5p | INSR | UAGCAGCACAUCAUGGUUUAC AAAAUUGACCAAUAGCUGCUGCUU .(((((((.(((((.))))).))))))). |  | −14.70 |
| miR-195-5p | UAGCAGCACAGAAAUAUUGGC AAAUUGACCAAUAGCUGCUGCUU .(((((((.((((((((.)).)))))).))))))). | 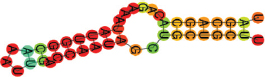 | −14.60 | |
| miR-27b-3p | UUCACAGUGGCUAAGUUCUGCCC UGCAGAGGAUUUAACUGUGAA .(((((((.((((((.)))))).))))))). | 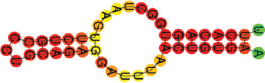 | −16.00 | |
| miR-128-3p | UCACAGUGAACCGGUCUCUUUCC UGCAGAGGAUUUAACUGUGAA (((((((.(((((.))))).))))))). | 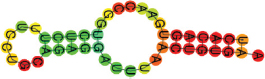 | −12.10 | |
| miR-379-5p | UGGUAGACUAUGGAACGUAGG UCUUUUUUCAGCACAGUCUACCU .((((((((.((.)).)))))))). | 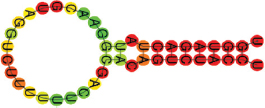 | −12.00 | |
| miR-424-5p | CAGCAGCAAUUCAUGUUUUG AAAAAUUGACCAAUAGCUGCUGCUU (((((((.(((.(((.))).))).))))))). | 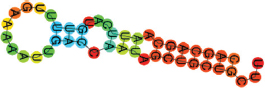 | −11.30 | |
| miR-29b-1-5p | GCUGGUUUCAUAUGGUGGUUU AGACACUGUAACAACAACAACCAGAA .((((((.(((((((.))))))).)))))). | 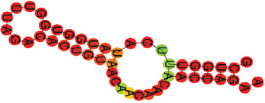 | −10.20 | |
| miR-15b-5p | IRS1 | UAGCAGCACAUCAUGGUUUA CAAAGAUACAUUUCAUCUGCUGCUG ((((((((.((.(((.((.)).))).)).)))))))) |  | −11.50 |
| miR-195-5p | UAGCAGCACAGAAAUAUUGG CAAGAUACAUUUCAUCUGCUGCUG ((((((((.((((((((.)))).)))).)))))))) | 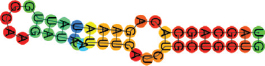 | −11.70 | |
| miR-27b-3p | UUCACAGUGGCUAAGUUCUGCG AAAAAAAAAAAUGCACUGUGAC .((((((((.)))))))). | 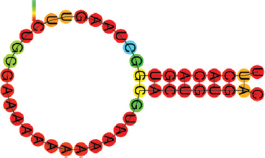 | −10.10 | |
| miR-128-3p | UCACAGUGAACCGGUCUCUUUCG AAAAAAAAAAAUGCACUGUGA ((((((((.(((.))).)))))))) |  | −9.00 | |
| miR-30d-5p | UGUAAACAUCCCCGACUGG AAGGCAUCCAUUUCAGUUUGUUUACU .(((((((.((((((((((.)).))))))))))))))). |  | −13.40 | |
| miR-143-5p | GGUGCAGUGCUGCAUCUCUGGUGA UUUCAUUGACUGAACUGCACG .(((((((.((.(((.))).)).))))))). |  | −11.90 | |
| miR-96-5p | UUUGGCACUAGCACAUUUUUGCUU GCACGUUCUAUAUUGUGCCAAG .(((((((.((((.)))).))))))). | 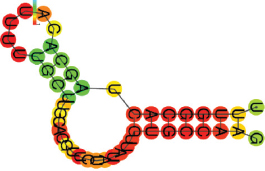 | −10.20 | |
| miR-144-3p | UACAGUAUAGAUGAUGUACUUU CAUAAUAAACUAGAUACUGUU .(((((((.((((.)))).))))))). |  | −6.30 | |
| miR-424-5p | CAGCAGCAAUUCAUGUUUUG AAAAGAUACAUUUCAUCUGCUGCUG ((((((((.((((.((.)).)))).)))))))) | 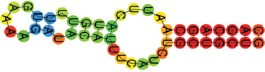 | −12.60 | |
| miR-7-5p | UGGAAGACUAGUGAUUUUGUUGUU GAAGAGGAAAUUAAAGUCUUCCA (((((((((.(((((((.((.)).))))))).))))))))) |  | −17.30 | |
MFE: Minimum free energy.