1. INTRODUCTION
Nowadays, the therapeutic efficacy of synthetic drugs is limited, because they can also adversely affect normal cells and tissues [1]. In this connection, various researchers in worldwide have been working toward developing effective anticancer medications from various sources, particularly plant based natural sources. Here, the wide varieties of herbs that has huge medicinal properties around the world have been found quite astonishing. Hence, herbal medicines are widely used in underdeveloped countries as an alternative medication to treat a number of diseases [2]. Followed by medicinal herbs contain several varieties of biochemical substances that were seen as effective in treating chronic and infectious diseases [3]. Various biochemical compounds such as alkaloids, amino acids, flavonoids, phenols, inorganic acids, tannins, terpenes, and carboxylic acids have been found in some of the medicinal plants [4]. In this connection, alkaloids are the secondary metabolites that have potent pharmacological activity. They are classified according to their biosynthetic precursors and heterocyclic compound structures. In general, indole alkaloids such as vincristine and vinblastine were employed for antineoplastic drugs isolated from Catharanthus roseus [5]. In addition, an indole alkaloid (aervine [10-Hydroxycanthin-6-one]) extracted from Aerva lanata (Amaranthaceae family) has antioxidant properties proved by Boobalan and Kamalanathan [6].
Here, Aerva javanica (Burm.f.) Schult was a seasonal medicinal herb in the Amaranthaceae family with rich in alkaloids and phenols and it has vertical branches that may reach heights of 5 to 6 feet. It was indigenous to tropical regions, particularly in Africa, India, Pakistan, and other Asian nations [7]. A. javanica was studied for its antibacterial activities [8], antidiarrheal, antiplasmodial and antiviral components [9], cytotoxic and cytogenetical [10], antioxidant [11], and anti-urolithiasis property [12]. Further, its roots are used to ease pain in kidney ailments and seeds as headache reliever. Hence, it was a seasonal plant that grows limitedly, and the pharmaceutical industry uses it extensively in medicines and health products. Establishing a successful transformation process was the first stage in improving the harvest value and product quality, which calls for genomic intercession.
Agrobacterium rhizogenes has been employed recently to produce hairy roots (HR), which has a suitable method due to their steady development and improved capacity to synthesize bioactive molecules in a hormone-free environment [13]. HR was triggered by A. rhizogenes, a soil bacterium which contains a large root-inducing plasmid (Ri plasmid) that fuses its t-DNA (transfer DNA) into the wounded site of plant genome [14]. The expression of role genes, such as rol A, B, C, and D, has been attributed for the HR development [15]. Recently, HRs have been used widely as biological matrices to produce essential metabolites due to their attractive characteristics, including high genetic stability and relatively fast growth. Due to the risks associated with harvesting roots for medicinal and chemical purposes, HR cultivation from various therapeutic plant species has increased [16].
Cancer was known as the second leading factor for mortality globally. Cancer affects millions of people every year across the globe and considered a dreadful, multifactorial disease characterized by causing abnormalities in genes, uncontrolled, and rapid division of cells [17]. Breast cancer is the second most malignant disease afflicting females worldwide. Exclusively, few environmental factors such as UV radiation, carcinogenic chemicals, and regular intake of fried junk foods such as grilled and barbeque chicken were the major causes of breast cancer [18]. Cancer treatment poses many challenges due to its complexity and makes it difficult to find therapeutic molecules for breast cancer treatment. Consequently, innovative treatment techniques are vital for treating the diseases. At present, clinical methods for treating cancers such as hormone therapy, radiotherapy, mastectomy, and chemotherapy are systemic anti-proliferative agents that distort cellular division and are in the use [19]. This needs much expertise, funds, time, and ethical issues behind. To overcome, these in silico methods can be employed for assessing the desired activity beforehand [20]. The computational tools are used to screen the active compounds from HRs of A. javanica by inspecting their active interactions with their respective target receptors. This work was continuation of current research problem on enhancement of indole alkaloids by HR cultures and focused on in silico and in vitro anticancer activity of aervine against breast cancer.
2. METHODOLOGY
2.1. Sample Collection
Young A. javanica plants were collected from the vicinity of Tiruchengode, Namakkal district, Tamil Nadu, India. The plant specimens were authenticated as A. javanica (Burm.f.) Juss. ex Schult by the Botanical Survey of India, Coimbatore (BSI/SRC/5/23/2020/Tech./726). Strains of A. rhizogenes MTCC 2364, MTCC 532, ATCC 15834, R1000, and LBA 9204 were procured from Department of Biotechnology, K. S. Rangasamy College of Technology (A). Cultures were maintained in glycerol (20% v/v) stock and the rate of viability was monitored employing regular subculture using LB broth.
2.2. Callus Induction
The leaves (whole leaf) and shoots (10–20 mm segments) were taken from 2-month-old wild A. javanica plant grown in the botanical garden located at the K. S. Rangasamy College of Arts and Science campus. The explants (leaf and shoot) were picked off and surface sterilization was done with running tap water for 5 min, followed by Bavistin (2% w/v) and Tween-20 (2% v/v) each for 60 s. Subsequently, the explants were again disinfected with sodium hypochlorite (2.50%) and mercuric chloride (0.1%) in the laminar air flow chamber and washed thoroughly with sterile distilled water for 5 times to eliminate trace chemicals. The adsorbed water on the surroundings of the explants was removed using disinfected filter paper before inoculation. The explants were inoculated on fresh MS semi-solid medium containing sucrose (3% w/v), agar PT (0.8% w/v), and plant growth regulators (PGRs) with combinations of 2, 4-D (0.5–2.5 mg/L) and NAA (0.5–2.5 mg/L). The inoculated explants were monitored twice a day and maintained at 24 ± 2°C under cool white fluorescent lights for 16 h and 8 h at dark. The callus induction frequency was recorded after 4 weeks of incubation [21].
2.3. A. rhizogenes Strains and Culture Condition
Five strains of A. rhizogenes, namely, ATCC 15834, R1000, LBA 9204, MTCC 2364, and MTCC 532 were revived from a glycerol stock and were used in genetic transformation experiments. The bacterial strains were grown individually in 100 mL LB (Luria-Bertani) broth containing rifampicin (50 mg/L, w/v), pH ≈ 7.2, and incubated at 28°C overnight in an orbital incubator shaker at 130 rpm until the cell suspension reached an optical density of 0.6 at 600 nm. Later, the overnight grown bacterial suspension was centrifuged at 4000 rpm for 10 min and the bacterial pellet was collected. It was re-suspended in the MS liquid medium (half strength) supplemented with acetosyringone (100 μM), kept in orbital shaker for 16–18 h, and used for bacterial transfection.
2.4. Cocultivation and Establishment of HR
For HR induction, germ-free young in vivo leaf (1 cm2), shoot (2 cm), and 4-week-old callus were aseptically injured with a sterile scalpel and immersed in the 16–18 h old bacterial culture (A. rhizogenes) containing (0.1 mg/L) acetosyringone and placed in the orbital shaker (80 rpm) for 20 min. The infected explants after washing 3 times with sterile distilled water were dried under an aseptic paper towel and inoculated on the freshly prepared 50% MS medium without antibiotics for 48 h at dark. After 48-h cocultivation, the explants were rinsed using sterile distilled water and implanted on the half-strength MS [22] medium with PT agar 0.8% (w/v), 500 mg/L antibiotics (Cefotaxime) devoid of PGRs was prepared, and pH was altered to 5.7 ± 0.5. Regular subculturing of explants was performed every week to avert browning and contamination. After 5 weeks of subculture, the HR from the three explants was harvested and mass propagated by cell suspension culture. Simultaneously, explants void of bacterial transformation was transferred to a solidified basal 50% MS medium incorporated with 0.5 mg/L of IAA and grown roots (non-transformed) were considered as control [23].
2.5.Genomic DNA Isolation and PCR Amplification
PCR was used to polymerize the total genomic DNA of the genetically transformed roots from A. javanica using modified protocols using CTAB (Cetyl Trimethyl Ammonium Bromide). DNA extracted from the roots of non-transformed A. javanica was used as the experimental negative control, whereas DNA from A. rhizogenes strain ATCC 15834 was used as the positive control. Primers of rol genes (rol B, 797 bp and rol C, 543 bp) used for PCR analysis; rol B (forward primer: 5′-CAA TGG ATC CCA AAT TGC TAT TCC-3′ and reverse primer: 5′-CGG CTT TAG GCT TCT TTC TTG AGG-3′) and rol C (forward primer: 5′-ATG GCT GAA GAC GAC CTG TGT-3′ and reverse primer 5′-TAG CCG ATT GCA AAC TTG CAC-3′). Further, vir D2 was recognized using 5′- ATG CCC GAT CGA GCT CAA GT-3′ and 5′-CCT GAC CCA AAC ATC TCG GCT-3′ primers and 338 bp fragments were amplified. PCR (Prima-96 plus, Thermal Cyclers) was carried out by the following procedures: amplification process of 32 cycles was undertaken (60 s denaturation at 94°C, 60 s annealing at 55°C, and 60 s extensions at 72°C). Before that, genomic DNA initial denaturation was set at 95°C for 300 s and the final extension was carried out for 420 s at 72°C. The amplified DNA was separated on agarose gel electrophoresis (1.5% w/v), ethidium bromide staining was used for visualization, and 1000 bp DNA ladder were used to identify the rol gene fragment [24].
2.6. Extract Preparation
All three HR samples were dried for 48 h in a circulating air oven at 37°C, after which the powders were milled and stored in dark flasks with slight modifications [11]. Thereafter, 10 g of each HRWL (HR from wild leaves), HRWS (HR from wild shoots), and HRIC (HR from in vitro callus) were submitted to successive extractions with chloroform by maceration in a ratio of 1:10 for the HR powder/solvent. The extracted solutions were filtered and concentrated in a rotary evaporator and kept in glass desiccators under vacuum and are used for UPLC separation.
2.7. UPLC Analysis
The chloroform extracts from mature HRs of A. javanica were analyzed for separation of biologically active compound as per the manufacturer’s procedure in Waters ACQUITY UPLC system (Waters, Milford, MA). Data acquisition and interpretation done by Empower 3.0 software in UHPLC-PDA detector (Waters Corp., Milford, MA) fixed with an Acquity UPLC BEH C18 column (2.1 × 100 mm, 1.7 μm; Waters Corp.) and 5 μL sample injection with a column temperature of 35°C. Linear gradient elution of the binary mobile phase was used with acetonitrile (A) and aqueous (B), at 0.5 mL/min flow rate {gradient change from solvent A to B, as follows (v/v): 10–30% (0–5 min), 30–60% (5–10 min), 60–90% (10–15 min), and 90% (15–20 min)}. The PDA-UV spectra ranging from 200 to 400 nm and a separate channel at 240 nm were tuned for the detection of bioactive metabolites [25]. Standards used such as vanillin and quinine were purchased from HiMedia, Mumbai, India, and aervine was obtained from Chem Faces (Wuhan, China).
2.8. Molecular Docking
Docking analysis was performed using the SDF format files of ligands retrieved from the PubChem database. The X-ray crystal structures of the receptors matrix metalloproteinase-16, epidermal growth factor receptor (EGFR), human EGFR 4, insulin-like growth factor 1 receptor, estrogen receptor-α (ER-α), human EGFR 3, phosphatidylinositol 3-kinase, progesterone receptor (PR), breast cancer 1 protein (BRCA-1), vascular endothelial growth factor, poly-ADP ribose polymerase 2, AKT (protein kinase B/PKB), and tumor necrosis factor-beta were retrieved from the Protein Data Bank having PDB IDs: 1RM8, 2J6M, 2R4B, 2ZM3, 3ERT, 3KEX, 4DNL, 4OAR, 4OFB, 4RN5, 5D5K, 6HHG, and 1TNR, respectively. Protein conformation screening was carried out on key targets using the RCSB PDB database (www.rcsb.org/). Small molecule ligands and water molecules were removed from the protein structure. Preparation of the proteins and compounds, polar hydrogen atoms, energy minimization, partial charges, and preparation of ligand was carried out using the Auto Dock Tools software, version 1.5.6. Simultaneously, a grid box was developed at the active cavities of the produced protein structure. Finally, all the docking was achieved using the Auto dock module available in PyRx software, version 0.8. The receptor-ligand pairs were sorted and screened based on affinity (kcal/mol) [26].
2.9. In Vitro Cytotoxicity Study MTT Assay
The MTT-based cytotoxicity assay was performed for the determination of the anti-proliferative activity of aervine enriched chloroform hairy root extract from in vitro callus (AECHRIC) to check the toxic effect on cell lines with slight modification [27]. Human cancer cell line MCF-7 (breast cancer) was grown in Dulbecco’s modified Eagle’s medium augmented with 10% fraction V (BSA) and 1× penicillin-streptomycin at 37°C and kept in a humidified chamber with 5% CO2. Tamoxifen was used as a reference drug (standard). Cells were suffused with 1 × 105 cells/well in a 96-well flat-bottom micro titer plate, a day before treatment and grown. AECHRIC stock (1.0 mg/mL) was made with 5% DMSO, and further, working solution (100 μg/mL) was prepared in serum-free culture media. Thereafter, the cell line was treated with the 200, 100, 50, 15, 10, 5, and 1 μg/mL doses of AECHRIC for 12, 24, 48, and 72 h. After the treatment periods, the medium was discarded and 0.15 mL MTT dye was added to 0.6 mL fresh medium. The 96-well plates were wrapped in aluminum foil and incubated at 37°C for 3 h. The medium containing MTT was substituted with glycine (0.11 mL) buffer and DMSO (0.89 mL) was added to suspend the formazan crystals. The absorbance was measured at 570 nm using an absorbance microplate reader (ELX 800).
2.10. Statistical Analysis
Each treatment comprised 20 explants and was replicated thrice. The total number of explants responded, the total number of HRs regenerated per explant and the percentage of transformation efficiency were reported after 3 weeks of culture. One-way analysis of variance and statistical significance test was used to determine the entire data and by Duncan’s multiple range tests (DMRT), mean and standard error values were calculated with a significance level of P < 0.05 (SPSS version 21; IBM).
3. RESULTS AND DISCUSSION
3.1. Callus Induction
Leaf and shoot explants of A. javanica were cultured in a combined MS medium fortified with 2, 4-D and NAA at 25°C in a sterile plant tissue culture chamber. By the end of the incubation period (4 weeks), a majority of the explants had formed callus on their leaf surfaces and shoot edges using varied hormone levels [Table 1]. Here, the leaf explants showed better response to regeneration of callus than shoot, which can be directly related to hormone concentrations [Figure 1a-d]. Similarly, Iris sanguinea callus regeneration was influenced by hormone concentrations and types of exogenous hormones [28]. Various explant types and PGRs have shown to have different roles in callus induction, determining an appropriate explant for callus regeneration to produce important therapeutic compounds from Cnidium officinale [29]. Davies [30] noted that callus proliferation could be rapidly accelerated with an increased amount of auxin and lower amount of cytokinin in the MS medium. Furthermore, the study on callus induction found that Nepeta binaloudensis Jamzad leaf explants had better callus regeneration than other explants [31]. Mei-Yin and Sani [32] reported strong callus formation by Vernonia amygdalina leaf explants grown in MS medium infused with 2,4-D (0.5–2.0 mg/L). In these experiments, fresh MS medium with 0.5 mg/L NAA 1.0 mg/L 2, 4-D) showed the maximum rate of callus regeneration. Induced calli were fragile and too soft to handle. Among the remaining hormone concentrations of NAA + 2, 4-D produces the most consistent callus regeneration, while, in some rare cases, no callus regeneration occurred. A moderate amount of callus regeneration was observed at the concentration of 1.0 mg/L for each hormone, 2, 4-D, and NAA. A. javanica elite leaf showed a maximum rate of callus regeneration cultured with 2.00 mg/L 2,4-D and 0.50 mg/L SPM [11].
Table 1: Callogenesis of Agrobacterium javanica leaf and shoot explants in different concentrations of plant growth regulators.
| Hormone (mg/L) | Leaf | Shoot | |||
|---|---|---|---|---|---|
| 2,4-D1 | NAA2 | Callus regeneration rate3 | Callus morphology | Callus regeneration rate | Callus morphology |
| 0.50 | 0.50 | + | Black and hard | - | - |
| 0.50 | 1.00 | + | Brown and fragile | - | - |
| 0.50 | 1.50 | ++ | Brown and spongy | + | Black hard |
| 0.50 | 2.00 | + | Pale brown and soft | + | Brown hard |
| 1.00 | 0.50 | ++++ | Pale brown and fragile | +++ | Pale brown fragile |
| 1.00 | 1.00 | ++ | Yellowish-white and fragile | ++ | Pale brown spongy |
| 1.00 | 1.50 | ++ | Pale yellow and fragile | + | Brown soft |
| 1.00 | 2.00 | + | Pale brown and fragile | + | Brown hard |
| 2.00 | 0.50 | + | Brown and fragile | - | - |
| 2.00 | 1.00 | + | Dark brown and hard | - | - |
| 2.00 | 1.50 | - | - | - | - |
1 2, 4 – D- 2, 4: dichlorophenoxyacetic acid;
2 NAA-1: naphthalene acetic acid,
3 +: Rate of callus development. + + + +: excellent, + + +: good, + +: moderate, + +: poor, - no callus development. Data was taken from triplicate experiments.
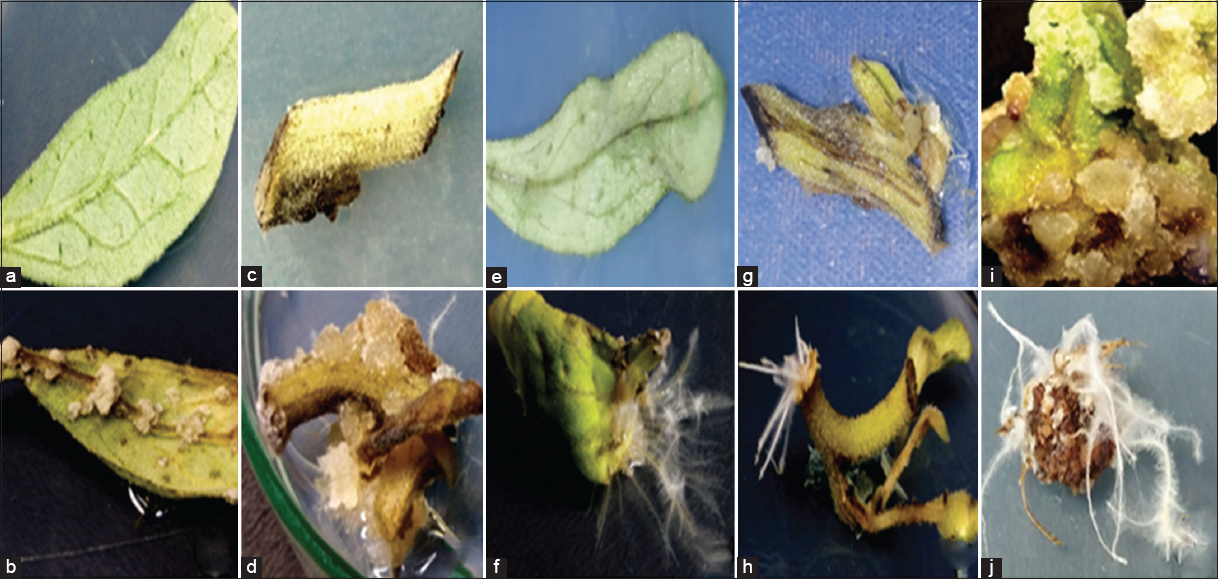 | Figure 1: Hairy root regeneration from different explants of Aerva javanica; (a) surface- sterilized leaf explants on MS medium prepared with the combination of 1.0 mg/L of 2.4-D and 0.5mg/L of NAA, (b) formation of callus on leaf explants after the completion of incubation time of 25 days at dark, (c) inoculated stem explants on MS callus medium, (d) soft spongy callus regenerated from shoot explants after 22–25 days of incubation with 2, 4 D (1.0 mg/L) and NAA (0.5 mg/L), (e) surface sterilized and ATCC15834 strain infected leaf explants kept for cocultivation at ½ strength MS medium without plant growth regulators at 24 ± 2°, (f) transformed root growth from leaf explants infected with Agrobacterium rhizogenes strain ATCC15834 on leaf explants (48 h of cocultivation), (g) A. rhizogens strain (ATCC15834) infected stem explant placed on ½ strength MS medium supplemented with antibiotic (250 mg/L of Cefotaxime), (h) Hairy root regeneration from shoots explants after completing cocultivation duration (5 days), (i) 25 days old callus culture from leaf explants placed on ½ MS medium without PGR after the infection with ATCC15834 strain, and (j) callus producing hairy roots after cocultivation with ATCC15834 strain (2 days). [Click here to view] |
3.2. Effect of A. rhizogenes Strains, Explants, and Infectivity on HR Induction
In this study, HR formation was directly influenced by different A. rhizogenes strains such as ATCC 15834, R1000, LBA 9204, MTCC 2364, and MTCC 532 infected for various time durations (0–25 min) with three explants (wild leaf, stem, and callus) of A. javanica [Figure 1e-j]. Transferred DNA (T-DNA) from the Ri (root-inducing) plasmid of the A. rhizogenes bacteria was integrated and expressed in the plant genome, resulting in HR induction [33]. Observing the hair like projections around the roots (similar to cat hairs) helped to identify the morphology of the HR formed by A. rhizogenes. As in this study, different A. rhizogenes strains were used to induce HR in C. roseus [34]. The five strains showed varying degrees of infection after 48 h of cocultivation. Similarly, Artemisia annua and Berberis aristata were also found effective in HR induction after a cocultivation time of 48 h [35,36]. It has been demonstrated that cocultivation time of less than 48 h reduces the rate of Ri plasmid transformation and over 48 h spoils the nature of the explants and affects the HR induction. Transformation frequency was strongly related to cocultivation duration and 2 days has been the most effective. In this case, tissue deterioration was observed when cocultivation lasted more than 2 days due to bacterial proliferation [37]. According to the infection period, all strains were significantly involved in generating HRs on in vitro calluses, wild shoots, and leaves.
According to Giri et al. [35], A. rhizogenes strains are different in their virulence, which leads to the different development rates of HR. A 20-min infection time was recommended by [38] for the transfer of the Ri plasmid to Portulaca oleracea. Strain ATCC 15834 had shown the highest transformation frequency (83 ± 5.2%) after 5 days of cocultivation [39]. Similarly, the ATCC 15834 strain exhibited high transformation efficiency on a variety of plants, such as Origanum vulgare, [40] Semecarpusana cardium, [41] and Artemisia aucheri [42]. Then, strain ATCC 15834 has proven superior in many cases to other strains. The strain (ATCC15834) was the most powerful in inducing HR, with highest transformation efficiency is denoted in Figure 2. Furthermore, during the HR induction, the 10, 15, and 25 min infection periods showed significantly lower infection rates towards all the three explants examined. At 0 and 5 min, none of these explants were infected. Due to inadequate bacterial infection duration, shorter infection periods (0, 5, 10, and 15 min) were not effective. A positive rate of infectivity was not found when the infection duration crossed the 20-min mark. All three explants (i.e., leaf, shoot, and callus) had the capacity of inducing HR after being infected with various strains of A. rhizogenes at different infection periods [Figure 2a-c]. Due to their strong meristematic activity, calluses showed the highest transformation compared to leaves and shoots. Therefore, 20 min was determined to be the optimal time for infection. A study showing results similar to our study used different strains, namely, ATCC15834, R1000, and C58, to induce HR in Trigonella foenum-graecum [43]. As in this experiment, after a 1-h infection period, callus was found to be most effective at triggering HR [36]. A. rhizogenes strains and types of explants have been found to exert a significant effect on the induction of HRs in Papaver bracteatum [23]. Transgenic root lines with the fastest growth rate and the strongest side-branching were selected for mass propagation. HR growth was not observed in the control group. Overall, these results indicated that A. rhizogenes infection conditions, strains, and explants could be optimized to effectively induce HR.
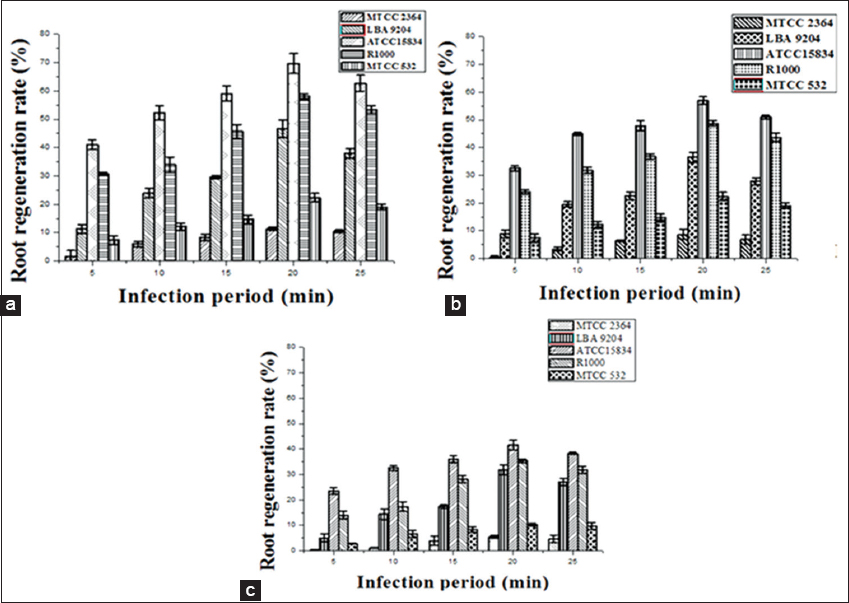 | Figure 2: Efficiency of Agrobacterium rhizogenes strains and incubation period on hairy root induction: (a) Root regeneration rate from callus explant based on infection period, (b) root regeneration rate from leaf explant based on infection period, and (c) root regeneration rate from shoot explant based on infection period. Data represented mean ± Standard Error of the three independent experiments. Mean values showed by different letters are significantly different at P < 0.05 (DMRT). [Click here to view] |
3.3. Transformation Confirmation by PCR Analysis
PCR-based identification confirmed the role of rol genes in the induction of HR from A. javanica with ATCC 15834. A. rhizogenes (ATCC 15834) induced HRs in plants, which served as template DNA for PCR analysis. In addition, genomic DNA from non-transformed roots was used as a negative control. The result of the PCR analysis of the regenerated roots showed the presence of rol B (650 bp) and rol C (480 bp), whereas no amplification response was observed in the non-transformed roots. Furthermore, rol B and rol C genes were successfully amplified in the root lines by PCR which was evident from the electrophoresis gel containing bands at 650 bp [Figure 3a; lanes 3, 4, 5, 6] and 480 bp [Figure 3b; lanes 3, 4, 5, 6]. No bands were observed in the lanes 4–6 of Figure 3c depicts that roots are not expressing vir D (370 bp) genes. Furthermore, no bands were seen at the experimental negative control [Figure 3a-c; lane 2]. The literature reports describe that transformation of plant tissues with rol A B C gene, as in this study, proliferates tissue growth inhibition due to the introduction of individual rol genes while sustaining increased secondary metabolite production [44]. According to Nilsson and Olsson, [45] the bacteria (A. rhizogenes) transfer two independent T-DNAs to the genomic plant (i.e., TL-DNA and TR-DNA). TL-DNA had the highest fundamental virulence factor and was required to produce HRs [46]. Amplification of the rol B and rol C genes has been done on different plants such as Picrorhiza kurroa [47] and Alpinia galangal [48]. Therefore, the PCR amplification of the rol B, rol C and vir D genes of A. javanica HR confirmed the involvement of A. rhizogenes in the transformation.
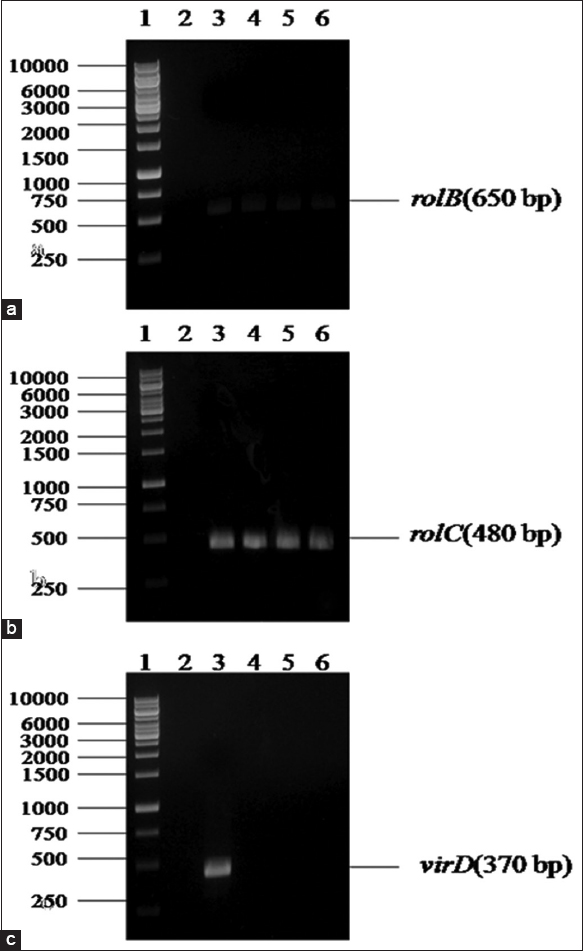 | Figure 3: PCR amplification of the rol B, rol C, and vir D genes from transformed and non- transformed root lines of Aerva javanica: (a and b) ane 1: DNA ladder 1000 bp. Lane 2: Experimental negative control (non-transformed roots). Lane 3: Positive control (Template DNA of Agrobacterium rhizogenes) strain ATCC15834. Lane 4: HR developed from wild leaves, Lane 5: HR developed from wild shoot and Lane 6: Hairy root lines regenerated from in vitro callus, and (c) Lane 1: DNA ladder 1000bp. Lane 2: Experimental negative control (non-transformed roots). Lane 3: Positive control (template DNA of A. rhizogenes) strain ATCC15834. Lane 4, 5, 6: No amplification of vir D gene occurred in HR developed from the wild leaf, wild shoot, and in vitro callus, respectively. [Click here to view] |
3.4. Cell Suspension Culture and Biomass Accumulation Based on Growth Time
In this study, the accumulation of bioactive components of HR produced by different explants (wild leaf, wild shoot, and in vitro callus) of A. javanica was determined based on the growth time. Almost all HR cultures accumulated bioactive components at different durations. After 50 days of cultivation, regeneration of HRs from the in vitro callus in the suspension culture demonstrated a greater capacity for mass propagation and bioactive component accumulation than roots regenerated from other explants. The HRIC cultures on the 42nd day accumulated 22.90 ± 0.78 g/L of fresh weight (FW) and 4.21 ± 0.44 g/L of dry weight (DW) biomass, which was approximately 30% higher than HRWL (16.97 ± 1.00 g/L FW and 3.27 ± 0.97 g/L DW) and approximately 45% than HRWS (13.18 ± 0.51 g/L FW and 2.57 ± 0.54 g/L DW) compared with other time intervals [Table 2]. Transformed root cultures of Panax quinquefolius acquired higher ginseng and saponin content levels than roots from wild plants [49]. Studies show that HR cultures accumulate more biologically active elements than plants grown in the field [50]. In the past, numerous reports have been published about HR regeneration and alkaloid (ajmalicine, catharanthine, serpentine, and vindoline) enhancement in C. roseus [51]. The phenolics and alkaloids have also been found to increase in HR cultures of Lobelia inflate L [52]. Therefore, it was concluded that, as a result of growth time, HRIC accumulated the maximum quantity of bioactive components.
Table 2: Bioactive component accumulation of hairy roots from different explants (wild leaf, shoot, and in vitro callus) of Aerva javanica.
| Time intervals (days) | HRWS1 | HRWL2 | HRIC3 | |||
|---|---|---|---|---|---|---|
| Fresh weight (g/L) | Dry weight (g/L) | Fresh weight (g/L) | Dry weight (g/L) | Fresh weight (g/L) | Dry weight (g/L) | |
| 00 | - | - | - | - | - | - |
| 07 | - | - | - | - | 5.09±0.12g | 1.05±0.12g |
| 14 | 3.08±0.91f | 0.64±0.20f | 6.47±0.46f | 1.23±0.24f | 8.13±0.21f | 1.67±0.71f |
| 21 | 6.01±0.31e | 1.01±0.85e | 8.82±0.70e | 1.66±0.10e | 11.67±1.10e | 2.08±0.15e |
| 28 | 7.92±0.97d | 1.59±0.23d | 12.6±1.11d | 2.14±0.48d | 15.25±0.63d | 3.19±0.80d |
| 35 | 10.38±0.86c | 2.07±0.82c | 15.21±1.46c | 3.09±0.37c | 19.57±0.54c | 3.95±1.05c |
| 42 | 13.18±0.51a | 2.57±0.54a | 16.97±1.00a | 3.27±0.97a | 22.90±0.78a | 4.21±0.44a |
| 50 | 12.07±0.60b | 2.19±0.43b | 15.86±1.22b | 3.00±0.92b | 21.87±2.03b | 4.08±0.43b |
1 HRWS: Hairy root from the wild shoot,
2 HRWL: Hairy root from wild leaf,
3 HRIC: Hairy root from in vitro callus. Table values are the mean±standard error of triplicates. Mean values followed by the different alphabets within a column vary significantly based on Duncan’s multiple range test (P < 0.05).
3.5. Comparison of Enhanced Bioactive Components from HR by UPLC Analysis
Enhancement of bioactive metabolites was detected using UPLC analysis and was based on retention time (RT). A total of 15 candidate metabolites were identified and three peaks were annotated by comparing their RTs with those of reference compounds. Despite this, 12 peaks remained unidentified. Surprisingly, one metabolite aervine was present at higher level, whereas two others vanillin and quinine were present at lower levels. According to our data, the higher abundance levels of three identified metabolites could have been directly influenced by the expression of role genes from A. rhizogenes (ATCC 15834). It was determined that compounds a, b, and c were aervine (RT = 0.661 min), vanillin (RT= 0.546 min), and quinine (RT = 2.632 min), respectively. Here, qualitative and quantitative analyses of bioactive metabolites were conducted for three samples (i.e., HRWL, HRWS, and HRIC). Compared to commercial standards (A-aervine, B-vanillin, and C-quinine) [Figure 4a and b], A. rhizogenes strain (ATCC 15834) showed higher aervine content in the HR regenerated from the wild stems, wild leaves, and in vitro callus. The HR culture enhanced a wide range of biologically active secondary metabolites that were beneficial to human populations. In all the samples, the compound aervine was detected at a significant level with a RT of 0.661 min, corresponding to the standard peak [Figure 4c-e].
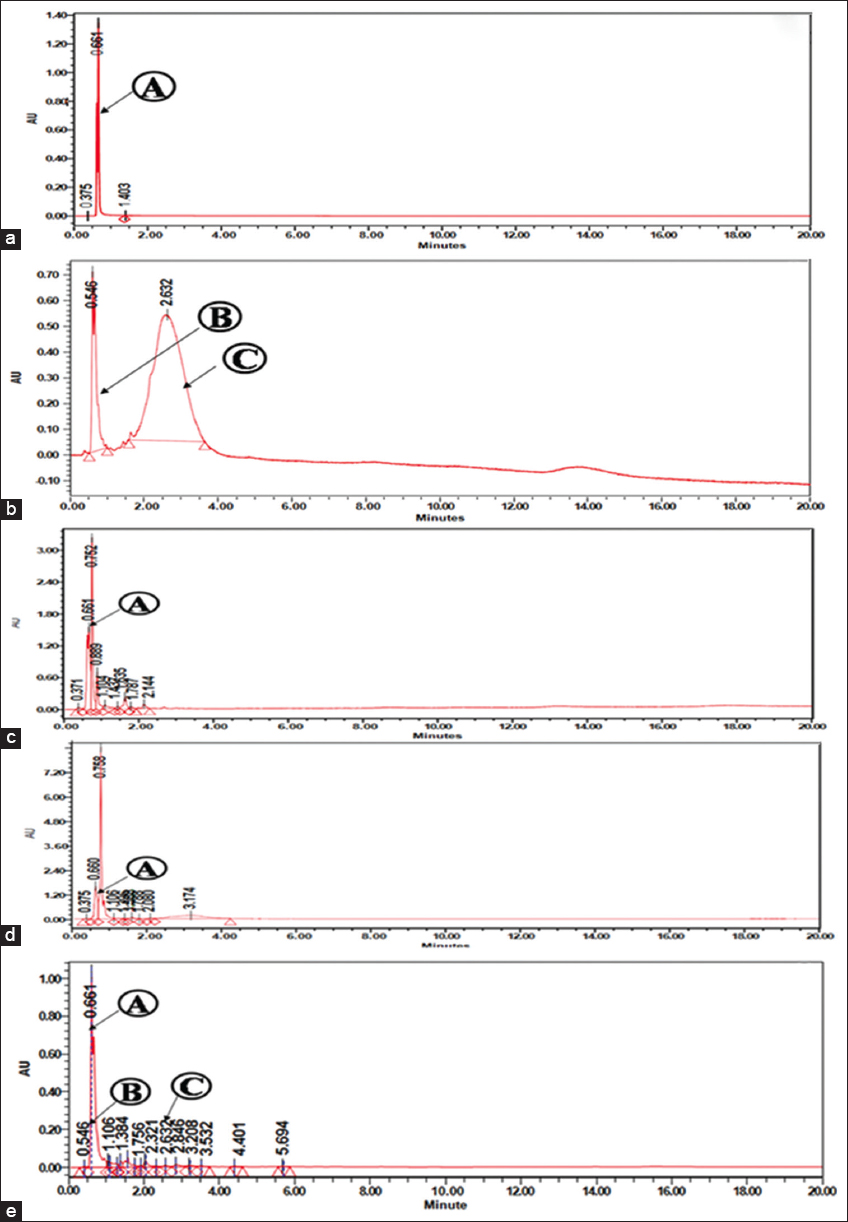 | Figure 4: TIC chromatogram of commercially available standards (a) Standard (A) aervine, (b) Standard (B) – vanillin, and (C) – quinine; TIC chromatogram of different bioactive compounds detected from chloroform extracts of Aerva javanica hairy roots from various explants: (c) hairy roots from the wild shoot (HRWS), (d) hairy roots from wild leaf (HRWL), and (e) hairy root from in vitro callus (HRIC). [Click here to view] |
Targeted/non-targeted bioactive components were determined using UPLC-qTOF [53,54]. Among the three samples analyzed, the amount of aervine in the HR from the callus produced the highest estimate (81.41 ± 2.82 μg/mL) with a RT of 0.661 min. Furthermore, HRWL (65.00 ± 3.67 μg/mL) and HRWS (53.06 ± 3.19 μg/mL) recorded moderate amount of aervine [Table 3]. Incorporating root-inducing plasmid from ATCC 15834 into callus explants caused a positive HR induction, which enhanced abundant bioactive components. Herewith, HRWS and HRWL accumulated a lesser amount of bioactive components due to the effect of A. rhizogenes (ATCC 15834). UHPLC-PDA was a conventional and validated method for quantifying and structurally identifying alkaloids without solvent extraction or compound isolation [55,56]. Similarly, in methanol extracts of opium [57], the alkaloids such as porphyroxine, morphine, codeine, noscapine, thebaine, and papaverine were separated and estimated using UHPLC-MS/MS. Through this analysis, it was found that the amount of aervine was three folds higher in the HRIC extract compared to HRWS and HRWL extracts.
Table 3: UPLC analysis data of enhanced bioactive components in hairy root of Aerva javanica.
| Name of the compound | Retention time (min) | Samples | ||
|---|---|---|---|---|
| HRWS1 | HRWL2 | HRIC3 | ||
| Aervine (µg/mL) | 0.661 | 42.92±2.97a | 55.00±3.67a | 71.41±2.82a |
| Quinine (µg/mL) | 0.546 | nd | nd | 38.69±0.94c |
| Vanillin (µg/mL) | 2.632 | nd | nd | 40.93±1.46b |
1 HRWS: Hairy root from the wild shoot,
2 HRWL: Hairy root from wild leaf,
3 HRIC: Hairy root form in vitro callus, nd: not detected. Table values are the mean±standard error of triplicates. Mean values followed by the different alphabets within a column are significantly varied based on Duncan’s multiple range test at (P < 0.05).
3.6. Establishment of in silico Simulation Studies to Predict the Binding Affinity and Orientations of Aervine with Different Breast Cancer Receptors
In silico docking is a preliminary tool to understand and gain the binding relationships between a small molecule (ligand) and its target protein receptor [58]. Due to the highest quantity of aervine in the HRIC extract, it was considered for in silico analysis. The binding modes of aervine against 13 different breast cancer target proteins to their active sites were determined by an in silico binding analysis using AutoDock, which is common and well-known docking software. The docking simulation study revealed that aervine could bind favorably to PR, ER-α, and BRCA-1 binding sites with the binding energy of −8.1, −7.5, and −7.0 kcal/mol, respectively, representing the towering binding affinity of the compound among the selected 13 target proteins. For the binding affinity for known inhibitor tamoxifen standard toward proteins, PR, ER-α, and BRCA-1 were docked [Table 4]. In a detailed binding interaction analysis, it was observed that aervine and tamoxifen participated in numerous important interactions with breast cancer target proteins, among which polar interactions were shown to be the majority. The hydroxyl and amide moiety from aervine on PR, ER-α and BRCA-1 were found to form hydrogen (H) bonds with Ser A: 728 (4.40 Å), Glu A: 353 (2.22 Å), and Leu A: 1701 (4.65 Å), respectively. Parallel to aervine binding, interaction was exposed by tamoxifen between some target proteins such as BRCA-1, in which the hydroxyl and amide moiety interacted with Lys B: 12 (5.71 Å) by hydrogen bond [Figure 5a and b].
Table 4: Binding affinities in kcal/mol for the test (Aervine) and standard (Tamoxifen) compound against protein targets of breast cancer.
| PDB Code1 | Target proteins | Binding affinities | |
|---|---|---|---|
| Aervine | Tamoxifen | ||
| 4OAR | PR (Progesterone receptor) | −8.1 | −7.2 |
| 3ERT | ER-α (Estrogen receptor-α) | −7.5 | −6.9 |
| 1RM8 | MMP (Matrix metalloproteinase-16) | −7.5 | −7.1 |
| 2R4B | HER4 (Human epidermal growth factor receptor 4) | −7.2 | −6.6 |
| 4OFB | BRCA-1 (Breast Cancer 1 protein) | −7.0 | −6.5 |
| 2ZM3 | IGFR (Insulin-like growth factor 1 receptor) | −7.3 | −7.4 |
| 3KEX | HER3 (Human epidermal growth factor receptor 3) | −7.1 | −7.0 |
| 5D5K | PARP (Poly-ADP ribose polymerase 2) | −6.9 | −7.3 |
| 6HHG | AKT (Protein kinase B/PKB) | −6.9 | −6.6 |
| 2J6M | EGFR (Epidermal growth factor receptor) | −6.8 | −6.6 |
| 4DNL | PI3K (Phosphatidylinositol 3-kinase) | −6.6 | −5.9 |
| 1TNR | TNF-β (Tumor necrosis Factor-beta) | −6.4 | −6.1 |
| 4RN5 | VEGF (Vascular endothelial growth factor) | −6.3 | −5.8 |
1 PDB: Protein data bank.
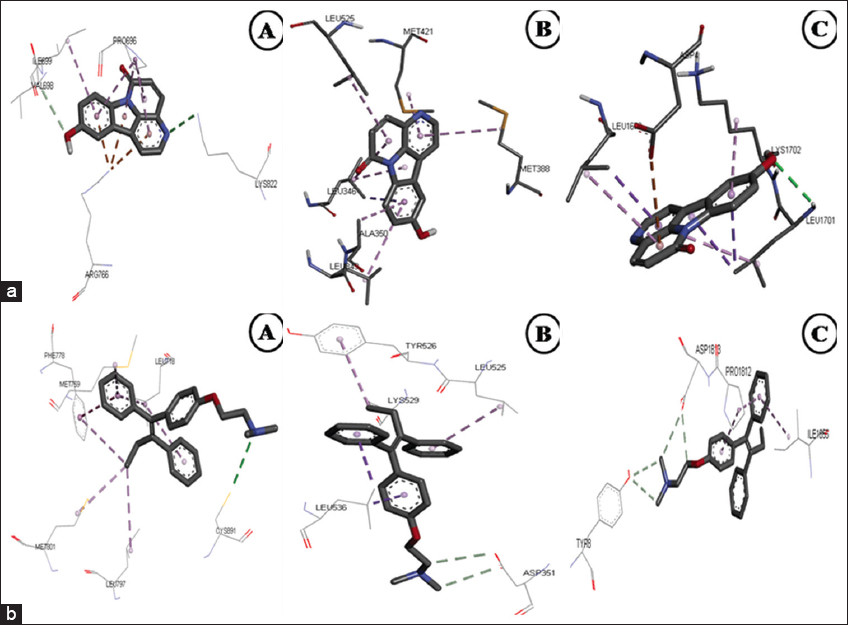 | Figure 5: (a) 3-D Docked confirmation of breast cancer target proteins with test compound aervine; (A) – PR (Progesterone receptor), (B) – ER-α (Estrogen receptor-α), and (C) – BRCA-1 (Breast Cancer 1 protein). (b) 3-D Docked confirmation of breast cancer target proteins with standard tamoxifen; (A) – PR (Progesterone receptor), (B) – ER-α (Estrogen receptor-α), and (C) – BRCA-1 (Breast Cancer 1 protein). [Click here to view] |
The results obtained in the present study was corroborated with the previous reports showing the active binding sites of similar breast cancer target proteins like MTOR representing similar amino acid patterns [59]. The compounds C-I and C-II show the binding affinity value of −9.4 and −7.5 kcal/mol, demonstrating that the isolated novel bioactive compounds from Nerium indicum can be a drug of option against cancer [20]. The aervine ligand molecule has a molecular weight of 236.22 amu (atomic mass unit). Likewise, the hydrogen donor was identified as 1 for aervine and 0 for tamoxifen; the hydrogen bond acceptor was found to be 4 for aervine and 2 for tamoxifen; and the value of logP was 2.14 for aervine and 6.00 for tamoxifen, respectively, and the molar refractivity was 69.68 for aervine and 119.72 for tamoxifen. The results of this study show that the isolated components from the HRs of A. javanica obey the regulations of Lipinski and could be a drug of preference to treat cancer.
3.7. Dose-dependent Cell Cytotoxicity Assay by MTT
Breast cancer (MCF-7) cell lines were exposed to AECHRIC extracts at various concentrations (200, 100, 50, 25, 10, and 1 μg/mL) and incubated for 72 h to determine cytotoxicity. It was found that AECHRIC extracts had a strong cytotoxic effect against breast cancer cell lines, depending on their concentration [Figure 6a-c]. Compared to the commercial standard tamoxifen, AECHRIC extract had a cytotoxic effect with an IC50 value of 149.86 ± 0.32 μg/ml, whereas tamoxifen had an IC50 value of 42.05 ± 0.92 μg/ml [Figure 6d-f]. The negative control exhibited no cytotoxicity (distilled water) [Figure 6g]. Similar studies were performed on the alkaloid portion of Terminalia acuminata to determine the effect of mitochondrial enzymes on cell growth inhibition by depleting soluble formazan crystals from MTT dye [60]. Fraction 17 from the MeOH extract from Clinacanthus nutans was most effective at inhibiting the proliferation of MDA-MB-231 and MCF-7 cancer cells [61]. According to the results of the research work of Abou Baker [62], a fraction made from Achillea millefolium extract was highly effective against MCF-7 cells, as measured by the IC50 value (0.87 ± 0.25 μg/mL). Thus, AECHRIC extracts may be more effective in cytotoxicity than commercial standards and require additional study in preclinical (animal models) and clinical trials against breast cancer in humans.
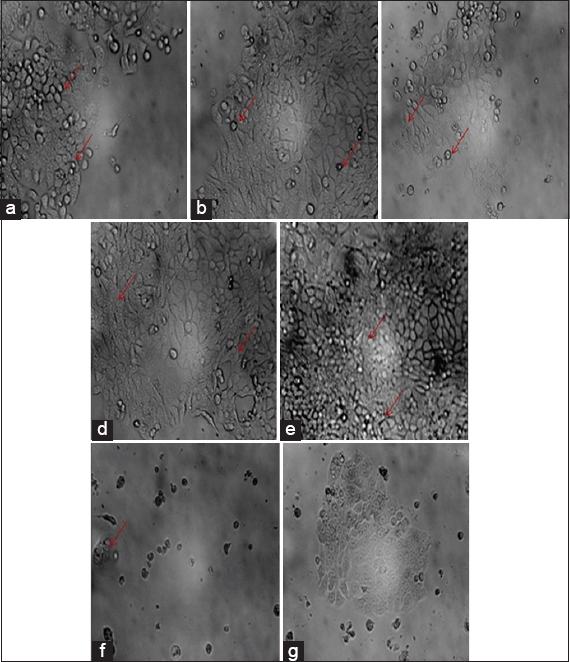 | Figure 6: Dose-dependent morphological changes of MCF-7 cell lines treated with aervine and tamoxifen for 72 h: (a) The microscopic images of MTT assays performed with 50 µg/mL of AECHRIC extract, (b) MTT colorimetric assay was performed using 100 µg/mL of AECHRIC extracts for 72 h at room temperature, (c) MTT dye absorbed by dead cells after treated with 200 µg/mL of AECHRIC extract, (d) MCF-7 cell lines treated with 50 µg/mL of standard after 72 h of treatment, (e) MTT dye absorbed by deceased cells after treated with 100 µg/mL of tamoxifen, (f) MTT dye absorption result after the MCF-7 cell lines treated with 200 µg/mL of standard (tamoxifen), and (g) negative control (Distilled water). NOTE-Arrows indicate apoptosis of MCF-7 Cell line. [Click here to view] |
4. CONCLUSION
In this present investigation, a rapid protocol for A. rhizogenes influenced hairy root culture in A. javanica for enhanced aervine production. The content of aervine in the HR regenerated from callus was three folds greater than the concentration observed in the leaf and shoot explants. The in silico and in vitro studies proved an efficient binding affinity of aervine with the active sites of the breast cancer target proteins. The use of hairy root culture in A. javanica could be an optional source of raw material in pharmaceutical industries to produce cancer drugs. Overall, aervine can be a promising drug against cancer that needs further validation through animal studies and this investigation would be an eye-opener for future research.
5. ACKNOWLEDGMENTS
The corresponding author and first author have indebted to express our sincere gratitude to the Department of Biotechnology, K. S. Rangasamy College of Arts and Science (Autonomous). This manuscript has been opted for preprint in Research Square: DOI: https://doi.org/10.21203/rs.3.rs-949387/v1.
6. AUTHORS’ CONTRIBUTIONS
RS, SB, DK: Conceptualization, analysis, data analysis, manuscript drafting, reviewing, and editing.
SM, VS, RS: UPLC analysis and in silico analysis.
7. FUNDING
No funding was received for conducting this study.
8. CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. AVAILABILITY OF DATA AND MATERIAL
The authors declare that the data supporting the findings of this study are represented as tables and charts, and all are available within the article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Alshatwi AA, Hasan TN, Shafi G, Syed NA, Al-Assaf AH, Alamri MS, et al. Validation of the antiproliferative effects of organic extracts from the green husk of Juglans regia L. on PC-3 human prostate cancer cells by assessment of apoptosis-related genes. Evid Based Complement Alternat Med 2012;2012:103026. [CrossRef]
2. Greenwell M, Rahman PK. Medicinal plants:Their use in anticancer treatment. Int J Pharm Sci Res 2015;6:4103-12.
3. Das NM, Sundari SS, Karuppusamy S, Mohan VR, Parthipan B. GC-MS analysis of leaf and stem bark of Cleidion nitidum (Muell-arg.) Thw. Ex Kurz. (Euphorbiaceae). Asian J Pharm Clin Res 2014;7:41-7.
4. Parekh J, Chanda V. In vitro antimicrobial activity and phytochemical analysis of some Indian medicinal plants. Turk J Biol 2007;31:53?8.
5. Costa-Lotufo LV, Montenegro RC, Alves AP, Madeira SV, Pessoa C, de Moraes ME, et al. The contribution of natural products as source of new anticancer drugs:Studies carried out at the national experimental oncology laboratory from the federal university of Ceara. Rev Virtual Quim 2010;2:47-58. [CrossRef]
6. Boobalan S, Kamalanathan D. Tailoring enhanced production of aervine in Aerva lanata (L.) Juss. Ex schult by Agrobacterium rhizogenes-mediated hairy root cultures. Ind Crops Prod 2020;155:112814. [CrossRef]
7. Samejo MQ, Memon S, Bhanger MI, Khan KM. Comparison of chemical composition of Aerva javanica seed essential oils obtained by different extraction methods. Pak J Pharm Sci 2013;26:757-60.
8. Sharif A, Ahmed E, Hussain MU, Malik A, Ashraf M. Antimicrobial constituents from Aerva javanica. J Chem Soc Pak 2011;33:439-23.
9. Ahmed EH, Nour BY, Mohammed YG, Khalid HS. Antiplasmodial activity of some medicinal plants used in Sudanese folk-medicine. Environ Health Insights 2010;4:1-6. [CrossRef]
10. Al-Fatimi M, Wurster M, Schroder G, Lindequist U. Antioxidant, antimicrobial and cytotoxic activities of selected medicinal plants from Yemen. J Ethnopharmacol 2007;111:657-66. [CrossRef]
11. Boobalan S, Kamalanathan D. Spermidine influences enhanced micropropagation and antibacterial activity in Aerva javanica (burm. F.) shult. Ind Crops Prod 2019;137:187-96. [CrossRef]
12. Ragini V, Kiran P. Anti urolithiatic activity of extract of Aerva javanica in rats. Int J Drug Dev Res 2014;6:35-45.
13. Tian L. Using hairy roots for production of valuable plant secondary metabolites. Adv Biochem Eng Biotechnol 2015;149:275-324. [CrossRef]
14. Chandra S. Natural plant genetic engineer Agrobacterium rhizogenes:Role of T-DNA in plant secondary metabolism. Biotechnol Lett 2012;34:407-15. [CrossRef]
15. Christey MC, Braun RH. Production of hairy root cultures and transgenic plants by Agrobacterium rhizogenes-mediated transformation. Methods Mol Biol 2005;286:47-60.
16. Zhou ML, Zhu XM, Shao JR, Tang YX, Wu YM. Production and metabolic engineering of bioactive substances in plant hairy root culture. Appl Microbiol Biotechnol 2011;90:1229-39. [CrossRef]
17. Parsa N. Environmental factors inducing human cancers. Iran J Public Health 2012;41:1-9. [CrossRef]
18. Marzbani B, Nazari J, Najafi F, Marzbani B, Shahabadi S, Amini M, et al. Dietary patterns, nutrition, and risk of breast cancer:A case-control study in the West of Iran. Epidemiol Health 2019;41:e2019003. [CrossRef]
19. Huang CY, Ju DT, Chang CF, Reddy PM, Velmurugan BK. A review on the effects of current chemotherapy drugs and natural agents in treating non-small cell lung cancer. Biomed (Taipei) 2017;7:23. [CrossRef]
20. Arunachalam T, Khader SZ, Ahmed SS, Vetrivel M, Ameen ST, Khadharu IS, et al. Radical scavenging and antiproliferative effect of novel phenolic derivatives isolated from Nerium indicum against human breast cancer cell line (MCF-7)-an in silico and in vitro approach. Environ Sci Pollut Res 2020;27:9038-57. [CrossRef]
21. Liao Q, Bai N, Wsman M, Sha H, Han HW, Liao ZL. Iris germanica L. Seedlings breeding technology research. Xinjiang Agric Sci 2017;54:470-8.
22. Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Plant Physiol 1962;15:473-97. [CrossRef]
23. Sharafi A, Sohi HH, Mousavi A, Azadi P, Razavi K, Ntui VO. A reliable and efficient protocol for inducing hairy roots in Papaver bracteatum. Plant Cell Tissue Organ Cult 2013;113:1-9. [CrossRef]
24. Tapia-Tussell R, Quijano-Ramayo A, Rojas-Herrera R, Larque-Saavedra A, Perez-Brito D. A fast, simple, and reliable high-yielding method for DNA extraction from different plant species. Mol Biotechnol 2005;31:137-9. [CrossRef]
25. Fu X, Cheng S, Liao Y, Huang B, Du B, Zeng W, et al. Comparative analysis of pigments in red and yellow banana fruit. Food Chem 2018;239:1009-18. [CrossRef]
26. Keerthanaa T, Boobalan S, Kamalanathan D, Karunakaran G, Sudha KG, Aarthi M, et al. Elicitation of apigenin in green leafy vegetable plants and its molecular docking evaluation for effective anticancer applications. Plant Cell Tissue Organ Cult 2022;150:459-78. [CrossRef]
27. Balupillai A, Nagarajan RP, Ramasamy K, Govindasamy K, Muthusamy G. Caffeic acid prevents UVB radiation induced photocarcinogenesis through regulation of PTEN signaling in human dermal fibroblasts and mouse skin. Toxicol Appl Pharmacol 2018;352:87-96. [CrossRef]
28. Liu H, Fan L, Song H, Tang B, Wang L. Establishment of regeneration system of callus pathway for Iris sanguine Donn ex Horn. In Vitro Cell Dev Biol 2020;56:694-702. [CrossRef]
29. Adil M, Ren X, Kang DI, Thi LT, Jeong BR. Effect of explant type and plant growth regulators on callus induction, growth and secondary metabolites production in Cnidium officinale Makino. Mol Biol Rep 2018;45:1919-27. [CrossRef]
30. Davies WP. Plant biotechnology:The genetic manipulation of plants. Slater A, Scotee N, Flower M 2003 Oxford:Oxford University Press. £19.99 (softback). Ann Bot 2004;94:646. [CrossRef]
31. Sagharyan M, Ganjeali A, Cheniany M, Kouhi SM. Optimization of callus induction with enhancing production of phenolic compounds production and antioxidants activity in callus cultures of Nepeta binaloudensis Jamzad (Lamiaceae). Iran J Biotechnol 2020;18:e2621.
32. Mei-Yin C, Sani H. In vitro Plantlet regeneration from nodal explant and callus induction of Vernonia amygdalina delile. J Plant Sci 2018;6:1-6.
33. Christensen B, Muller R. The use of Agrobacterium rhizogenes and its rol-genes for quality improvement in ornamentals. Eur J Hortic Sci 2009;74:275-87.
34. Toivonen L, Balsevich J, Kurz WG. Indole alkaloid production by hairy root cultures of Catharanthus roseus. Plant Cell Tissue Organ Cult 1989;18:79-93. [CrossRef]
35. Giri A, Giri CC, Dhingra V, Narasu ML. Enhanced podophyllotoxin production from Agrobacterium rhizogenes transformed cultures of Podophyllum hexandrum. Nat Prod Lett 2001;15:229-35. [CrossRef]
36. Brijwal L, Tamta S. Agrobacterium rhizogenes mediated hairy root induction in endangered Berberis aristata DC. Springerplus 2015;4:443. [CrossRef]
37. Sarkar J, Misra A, Banerjee N. Genetic transfection, hairy root induction and solasodine accumulation in elicited hairy root clone of Solanum erianthum D. Don. J Biotechnol 2020;323:238-45. [CrossRef]
38. Moghadam AY, Piri K, Bahramnejad B, Ghiasvand T. Dopamine production in hairy root cultures of Portulaca oleracea (Purslane) using Agrobacterium rhizogenes. J Agric Sci Technol 2014;16:409-20.
39. Tavassoli P, Afshar AS. Influence of different Agrobacterium rhizogenes strains on hairy root induction and analysis of phenolic and flavonoid compounds in marshmallow (Althaea officinalis L.). 3 Biotech 2018;8:351. [CrossRef]
40. Habibi P, de Sa MF, da Silva AL, Makhzoum A, da Luz Costa J, Borghetti IA, et al. Efficient genetic transformation and regeneration system from hairy root of Origanum vulgare. Physiol Mol Biol Plant 2016;22:271-7. [CrossRef]
41. Panda BM, Mehta UJ, Hazra S. Optimizing culture conditions for establishment of hairy root culture of Semecarpus anacardium L. 3 Biotech 2017;7:21. [CrossRef]
42. Sharafi A, Sohi HH, Mirzaee H, Azadi P. In vitro regeneration and Agrobacterium mediated genetic transformation of Artemisia aucheri boiss. Physiol Mol Biol Plants 2014;20:487-94. [CrossRef]
43. Zolfaghari F, Rashidi-Monfared S, Moieni A, Abedini D, Ebrahimi A. Improving diosgenin production and its biosynthesis in Trigonella foenum-graecum L. Hairy root cultures. Ind Crops Prod 2020;145:112075. [CrossRef]
44. Shkryl YN, Veremeichik GN, Bulgakov VP, Tchernoded GK, Mischenko NP, Fedoreyev SA, et al. Individual and combined effects of the rolA, B, and C genes on anthraquinone production in Rubia cordifolia transformed calli. Biotechnol Bioeng 2008;100:118-25. [CrossRef]
45. Nilsson O, Olsson O. Getting to the root:The role of the Agrobacterium rhizogenes rol genes in the formation of hairy roots. Physiol Plant 1997;100:463-73. [CrossRef]
46. Sujatha G, Korac-Zdravkovic S, Calic D, Flamini G, Kumari BD. High-efficiency Agrobacterium rhizogenes-mediated genetic transformation in Artemisia vulgaris:Hairy root production and essential oil analysis. Ind Crops Prod 2013;44:643-52. [CrossRef]
47. Verma PC, Singh H, Negi AS, Saxena G, Rahman LU, Banerjee S. Yield enhancement strategies for the production of picroliv from hairy root culture of Picrorhiza kurroa Royle ex benth. Plant Signal Behav 2015;10:e1023976. [CrossRef]
48. Rao K, Chodisetti B, Mangamoori LN, Giri A. Agrobacterium-mediated transformation in Alpinia galanga (Linn.) Willd. For enhanced acetoxychavicol acetate production. Appl Biochem Biotechnol 2012;168:339-47. [CrossRef]
49. Kochan E, Szymczyk P, Kuzma L, Szymanska G. Nitrogen and phosphorus as the factors affecting ginsenoside production in hairy root cultures of Panax quinquefolium cultivated in shake flasks and nutrient sprinkle bioreactor. Acta Physiol Plant 2016;38:1-149. [CrossRef]
50. Bathoju G, Rao K, Giri A. Production of sapogenins (stigmasterol and hecogenin) from genetically transformed hairy root cultures of Chlorophytum borivilianum (Safed musli). Plant Cell Tissue Organ Cult 2017;131:369-76. [CrossRef]
51. Almagro L, Fernandez-Perez F, Pedreno MA. Indole alkaloids from Catharanthus roseus:Bio production and their effect on human health. Molecules 2015;20:2973-3000. [CrossRef]
52. Balvanyos I, KursinszkiL, Banyai P, Szoke E. Analysis of polyacetylenes by HPLC in hairy root cultures of Lobelia inflate cultivated in bioreactor. Chromatographia 2004;60:235-8. [CrossRef]
53. Lopez-Cobo A, Gomez-Caravaca AM, Cerretani L, Segura-Carretero A, Fernandez-Gutierrez A. Distribution of phenolic compounds and other polar compounds in the tuber of Solanum tuberosum L. by HPLC-DAD-q-TOF and study of their antioxidant activity. J Food Compost Anal 2014;36:1-11. [CrossRef]
54. Talhaoui N, Gomez-Caravaca AM, Leon L, De la Rosa R, Segura-Carretero A, Fernandez-Gutierrez A. Determination of phenolic compounds of 'Sikitita'olive leaves by HPLC-DAD-TOF-MS. Comparison with its parents 'Arbequina'and 'Picual'olive leaves. LWT Food Sci Technol 2014;58:28-34. [CrossRef]
55. Lurie IS, Driscoll SE, Cathapermal SS, Panicker S. Determination of heroin and basic impurities for drug profiling by ultra-high-pressure liquid chromatography. Forensic Sci Int 2013;231:300-5. [CrossRef]
56. Hubert J, Nuzillard JM, Renault JH. Dereplication strategies in natural product research:How many tools and methodologies behind the same concept?Phytochem Rev 2017;16:55-95. [CrossRef]
57. Li L, Panicker S, Casale EM. UHPLC-MS/MS quantitation of porphyroxine in opium and application of porphyroxine-acetylated products as signature markers for heroin. Drug Test Anal 2019;11:999-1008. [CrossRef]
58. Alonso H, Bliznyuk AA, Gready JE. Combining docking and molecular dynamic simulations in drug design. Med Res Rev 2006;26:531-68. [CrossRef]
59. Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBPI 2-rapamycin complex interacting with the binding domain of human FRAP. Science 1996;273:239-42. [CrossRef]
60. Senthilraja P, Kathiresan K. In vitro cytotoxicity MTT assay in vero, HepG2 and MCF-7 cell lines study of Marine yeast. J Appl Pharm Sci 2015;5:80-4. [CrossRef]
61. Mutazah R, Hamid HA, Ramli AN, Aluwi MF, Yusoff MM. In vitro cytotoxicity of Clinacanthus nutans fractions on breast cancer cells and molecular docking study of sulphur containing compounds against caspase-3. Food Chem Toxicol 2020;135:110869. [CrossRef]
62. Baker DH. Achillea millefolium L. Ethyl acetate fraction induces apoptosis and cell cycle arrest in human cervical cancer (HeLa) cells. Ann Agric Sci 2020;65:42-8. [CrossRef]