1. INTRODUCTION
Arachis hypogaea L, commonly known as groundnut is one of the most cultivated plants in Indonesia. Yet, its waste utilization is still relatively low. According to the Central Bureau of Statistics in 2018, groundnut production in Indonesia reached 512,198 tons/year with groundnut shell weights approximately 29% of the total weight of its pod. Therefore, the estimated amount of groundnut shell in Indonesia that has not been valorized reaches 148,537 tons/year [1]. Groundnut shell contains a relatively high amount of cellulose (35.7%), hemicellulose (18.7%), lignin (30.2%), and ash (5.9%) [2]. Groundnut shell is also known as a source of antioxidants as well as raw materials for fertilizers and production of cellulase and hemicellulose enzymes [3].
Under natural conditions, the hydrolysis of cellulose occurs with the help of cellulase, a’ combination of three groups of enzymes particularly endogluconase (1,4-β-D-glucanohydrolase, EC 3.2.1.4), exogluconase (1,4-β-D-glucan selobiohydrolase, EC 3.2.1.91), and β-glucosidase (β-D-glucoside glucohydrolase, EC 3.2.1.21). Endoglucanase attacks O-glycosidic bonds randomly and produces glucan chain of different lengths wheres exoglucanase attacks the end of cellulose chain and produces cellobiose as the final product while β-Glucosidase hydrolyzes the terminal part of the cellulose chain and yields β-D-glucose [4].
Cellulase is typically synthesized by various microorganisms such as fungi and bacteria with lignocellulose as a substrate. Several genera of microbes that have been most studied and known as the best biological agents in producing cellulase are Clostridium, Cellulomonas, Thermomonospora, Trchoderma, and Aspergillus [5,6]. Trichoderma is one of the best-known fungal genera in producing cellulase. Enzymatic activity yields from this genus can be increased by co-culturing Trichoderma with organisms that support the synthesis of cellulase, such as Aspergillus and Rhizopus which can be isolated from tape and tempeh yeast. Mixed cultures of these two genuses and Trichoderma can produce cellulase with a relatively high enzymatic activity [7].
The best-known fermentation method for producing cellulase enzymes is the Solid-State Fermentation (SSF) method that uses a solid substrate as a medium for the culture of filamentous fungi. Enzymes are produced on substrate surfaces with low or no moisture content. Based on previous studies, it is known that SSF method can provide a higher enzyme yield than the Submerged Fermentation (SMF) method [8]. SSF method can maximize contact between microorganism and the surface of the substrate, so that microbes have an abundant source of nutrients for the fermentation process to be carried out. There are many factors that must be considered in the fermentation process of SSF. They are substrate, moisture content, temperature, pH, concentration of additional macro and micronutrients, and the fermentation time. The exact values of these parameters will depend on the biological agents and the main substrate used in the fermentation as well as the desired products [9].
At present, systematic studies that have reported on the SSF of groundnut shell using mixed culture of mixed culture of Trichoderma sp., tape yeast, and tempeh yeast to produce cellulase are very scarce. Hence, this study was carried out to valorize groundnut shell as a substrate for SSF using mixed culture of Trichoderma sp., tape yeast, and tempeh yeast to produce cellulase. Mathematical models were also developed to estimate relevant kinetic parameters in SSF and SMF to produce cellulase.
2. MATERIALS AND METHODS
2.1. Materials
Groundnut shell was obtained from Surakarta, Central Java, Indonesia. A commercial culture of bio fungicide consisting of a mixture of Trichoderma harzianum, Trichoderma viride, Trichoderma sp., and rice as the carrier was purchased from the Indotani Lestari store. Commercial tempeh yeast (RAPRIMA) that consists of Rhizopus oligosporus. and tape yeast (CAP TAMPIR) that consists of Aspergillus sp., were purchased from online shops in Bandung. Phosphate, hydrochloric acid, sulfuric acid, sodium hydroxide, dinitrosalicylic acid (DNS), Rochelle salt, citric acid, acetic acid, nitric acid, and sodium citrate dihydrate were obtained from School of Life Sciences and Technology, Institut Teknologi Bandung (ITB), West Java, Indonesia.
2.2. Determination of Lignocellulose Composition in Groundnut Shell
Approximately, 1 g (weight a) sample of groundnut shell was dried. After drying, the sample was added with 150 ml of distilled water/benzene alcohol and then refluxed at 100°C using a water bath for 1 h. The reflux products were filtered and rinsed with 300 ml of hot water. The residue obtained from the filtering process was dried using an oven until a constant weight was obtained (weight b). The dried residue was added with 150 ml of 1 N H2SO4 and refluxed again using a water bath at 100°C for 1 h. After that, the reflux products were filtered, and the residue was rinsed with distilled water until the pH was neutral. The residue was then dried using an oven until a constant weight was obtained (weight c). The dried residue was then added with 100 ml of 72% H2SO4 and then soaked at room temperature (27°C) for 4 h. Next, the residue was refiltered. The filtered residue was added with 150 ml of 1 N H2SO4 and then refluxed using a water bath at 100°C for 1 h. After that, the reflux products were filtered and then rinsed with distilled water until the pH reached a neutral point. The residue was then dried using an oven at 105°C until a constant weight was obtained (weight d). The dried residue was then ashed using a furnace and weighed (weight e) [10]. The composition of cellulose, hemicellulose, and lignin content of the groundnut shell can be calculated using the following equations:
2.3. Solid-State Fermentation
Approximately, 5 g of groundnut shell mixed with 5 ml of water and 0.5 g of culture [11] particularly Trichoderma sp., tape yeast; tempeh yeast, and mixture of Trichoderma sp. with both yeast with a ratio of 1:1 (g/g). The fermentation was carried out in beaker glasses at room temperature (27°C), and sampling was carried out every 24 h in duplicate.
2.4. Determination of Fungal Biomass
Approximately, 1 g (weight a) of the mixture was dried using an oven until a constant weight was obtained. After drying, the mixture was weighed (weight b). The dried solids were then soaked in 5 ml of acetic acid reagent and boiled for 30 min using a water bath. Acetic acid reagent consists of mixture of acetic acid and nitric acid with ratio of 10:1 (v/v) and then cooled and centrifuged at 5500 rpm for 15 min. The solid phase obtained was then rinsed with distilled water and dried at 70°C using an oven until a constant weight was obtained (weight c) [12]. The dry weight (DW) of the fungal biomass can be calculated by equation (4).
2.5. Determination of Cellulase Activity (FPase)
After the fermentation process, extraction was carried out to separate the extract of a crude enzyme from the substrate by mixing the fermentation medium with phosphate buffer (pH 5) with a substrate to buffer ratio of 1:10 (weight/volume). The extraction was carried out at room temperature (27°C) using a shaker set at a speed of 120 rpm for 1 h [13] followed by filtration through a cloth filter. The filtrate obtained was centrifuged at 550 rpm for 20 min, and the resulting supernatant was separated for enzyme activity assay. Approximately, 0.5 ml of the crude enzyme was mixed with 1 ml of Na-citrate (pH 4.8) in a test tube. The test tube was immersed in a water bath at 50°C and incubated for 60 min. After that, 3 ml of DNS reagent was added to the tube. Test tube was then heated in a boiling water bath for 5 min followed with cooling by immersion in it water bath filled with water at room temperature (27°C). After that, 20 ml of distilled water was poured into the test tube and inverted several times until the mixture was homogenous. After that, the tube was settled down for at least 20 min, and the absorbance of the solution was measured at 540 nm [14]. One unit of enzyme activity is the required amount of enzyme to release 1 mol of reducing sugar per min per ml which can be calculated by equation (5).
where MW is a molecular weight of glucose (180 g/mol), t is incubation time (min), H is total volume of enzyme-substrate (ml), and E is volume of enzyme (ml).
All the analyses were performed in triplicate and expressed as mean ± standard deviation. Differences were tested with one-way analysis of variance (ANNOVA) followed by Duncan test using MINITAB 17.
2.7. Estimation of Kinetic Parameters
Modified Monod and Luedeking-Piret equations were used to determine relevant kinetic parameters, particularly ∞m, Ks, kd, Yx/s, ms, k1, k2, dan Ki. for modeling cell growth, enzymatic activity, and consumption of substrate using equation (6), (7) and (8), respectively [15].
where X is the total cell concentration of fungi in the medium (g/l), μm is the maximum growth rate (1/h), S is the concentration of substrate (g/l), Ks is the saturation constant of substrate (g/l), and kd is the constant for cell death (1/h).
where k1 is the synthesis rate constant for cellulase (IU/mL h), k2 is the decomposition rate constant for cellulase (1/h), and Ki is the coefficient for substrate inhibition.
where Yx/s is the yield of cell biomass per substrate (g/g), and ms is the specific maintenance coefficient (1/h). The equations were solved using ode113 function in MATLAB and compared with secondary data obtained from the literature for SSF and SmF (Castillo et al., 1994; Ma et al., 2013)
3. RESULTS AND DISCUSSION
3.1. Characterization of Lignocellulose Content in Groundnut Shell
Lignocellulose content of groundnut shell used in this study was determined using a Chesson-Datta method, and the results are shown in Table 1. Based on Table 1, the groundnut shell used in this study has a very low moisture content and primarily composed of hemicellulose; 50.15 ± 0.04% dw, followed by lignin; 33.70 ± 1.44% dw and cellulose; 12.24 ± 0.78% dw. The cellulose content obtained in this study is lower than the values reported in previous studies that can reach as high as 63.5% [16].
The differences may be due to the origin of the sample where the sample used in this study was obtained from Central Java, Indonesia meanwhile the values reported by other studies were obtained from different countries, particularly Japan, China, and Singapore [22,23]. Different cultivation locations may cause differences in soil temperature [24], altitude [21], light intensity [25], and soil moisture content [26,27] resulting in differences of lignocellulose content in the same varieties. Previous studies also have reported that drought stress has a great influence on the cellulose and hemicellulose content in biomass [27-30].
 | Table 1: Lignocellulose composition of groundnut shell. [Click here to view] |
3.2. Effect of Fermentation Time on Enzymatic Activity of Cellulase
Fermentation was carried out for 5 days, and the enzymatic activity of cellulase was determined daily and the results are shown in Figure 1. The enzymatic activity of cellulase observed in this study lies in the range of 0.06–0.12 filter paper unit (FPU)/ml. The highest activity of cellulase was obtained from the fermentation of Trichoderma sp and tape yeast that reaches up to 0.12 FPU/ml. Mixed cultures have a higher cellulase activity as compared to the single cultures, probably due to growth symbiosis in mixed cultures [3] and synergism of enzyme complexes [31,32] which may occur due to the presence of 3 different enzymes that work synergistically, endo-β-glucanase, exo-β-glucanase, and β-glucosidase [4]. It has been reported that Trichoderma sp typically produces cellulase with high levels of endoglucanase and exoglucanase activity, but relatively low β-glucosidase activity [7].
The difference in activity of the enzyme complex causes the enzyme complex to interact synergistically and produces a higher activity compared to single culture enzymes. In addition, the growth of microorganism in mixed cultures mat performed better than in single cultures due to the synergism of metabolites produced by both cultures can support the growth of both cultures [33]. Higher biomass growth of microorganisms can support the production of cellulase enzymes with more optimum activity. Cellulase activity may vary depending on the microorganism, substrate, fermentation time and fermentation method as shown in Table 2. From the table, it can be noted that the cellulase activity determined in this study and the literature is quite different. Differences in enzyme activity can be caused by several factors, such as provision of pre-treatment, and cellulose content in the substrate. Pre-treatment of the substrate before the fermentation is highly recommended. Physical pre-treatment in the form of steam explosion [37,38] and ultrasonic [39] or chemical treatment in the form of alkali [40] is known to increase cellulase enzyme activity in the fermentation process. In this study, the groundnut shells used for the fermentation process were not pretreated. Meanwhile, in the study of Mojsov [35] and Afriyanti et al. [36], the fermentation substrate was pretreated. This could be the reason why the enzyme activity in both studies was higher than this study.
Pre-treatment of the fermented substrate is known to help loosen the lignocellulose structure, increase the solubility of hemicellulose, reduce the crystal structure, and increase the surface area and pore on the substrate so that the enzyme hydrolysis process becomes easier [41]. The high content of lignin can inhibit the hydrolysis of cellulose. In addition to protecting the cellulose structure from being hydrolyzed by the cellulase enzyme, lignin is also known to be bound to the cellulase enzyme so that the activity of the cellulase enzyme decreases [42]. Pre-treatment with H2O2 [43] or lignin-decomposing fungi [44] is known to dissolve lignin compounds in lignocellulose so that the activity of cellulase enzymes can be increased. Pre-treatment on the substrate is known to increase the activity of the cellulase enzyme in the fermentation process from 1.2 to 6.9 times compared to the fermentation process without pre-treatment [3,39].
 | Figure 1: Maximum enzyme activity of 5 days fermentation at room temperature condition. [Click here to view] |
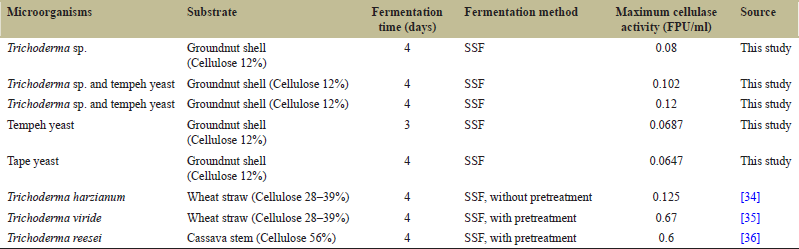 | Table 2: Comparison of cellulase activity using different substrate. [Click here to view] |
The cellulose content in the substrate can also affect the activity of the cellulase enzyme. Cellulase enzyme production by Trichoderma sp. is induced by the presence of inducer compounds such as pure cellulose, lactose, sophorose, and cellobiose [45,46]. Sophorose and cellobiose are compounds derived from cellulose [47]. Cellulase production by Trichoderma is also influenced by the presence of an inducer compound. The presence of the induction compound in the medium will be directly proportional to the production of cellulase. Because cellulase-inducing compounds are cellulose-derived compounds, the production of cellulase enzymes will be higher if the cellulose content in the substrate is also higher [45]. In this study, the groundnut shell substrate used has a cellulose content that is relatively small; 12.24 ± 0.78% dw. This could be the cause of low enzyme activity produced compared to Toscano et al. [34] that used a substrate with a higher cellulose content.
Nutrient source is one of the most important factors to produce metabolites. In this study, no additional nutrient sources were used, such as basal medium or Mendel and Reese’s medium which provided a source of nitrogen and vitamins for microorganisms. This simple medium is used by microbes as an early energy source in the initial condition to provide biomass growth. The nutrients obtained by microbes in this study were only provided by groundnut shells and carrier media, such as talc. This could be one of the causes of the lower cellulase activity. Adequate nutritional composition can maintain microbial growth so that microbes can grow optimally and produce cellulase activity more effectively. This study did not use an additional medium aimed at examining the production ability of Trichoderma sp and tempeh yeast using the lignocellulosic substrate as the only nutrient. On the other hand, the use of additional chemical mediums can increase production costs on a large scale. The cellulase production using lignocellulose as the sole nutrient can reduce production costs and reduce waste in the surrounding environment [48].
3.3. Effect of Fermentation Time on the Amount of Biomass
In this study, the growth of fungal biomass was calculated to determine the effect of different cultures on fungal growth. Fungal biomass was measured every 24 h for 5 days of fermentation. The result obtained from the fungal biomass measurement was tested using one-way ANOVA followed by Duncan post hoc test to analyze the results statistically. From the test, it can be concluded that fungal growth between each culture variations is significantly different (P < 0.05). The weight profile of fungal biomass throughout the fermentation period is presented in Figure 2.
From the figure, the maximum weight of fungal biomass was obtained from mixed culture of Trichoderma sp. and tempeh yeast with a biomass weight up to 0.34 ± 0.00 g. That value was significantly higher than the others. This could be due to the synergism between the metabolites released by Trichoderma sp and tempeh yeast which mutually support the metabolic processes of the microorganisms [33]. However, Trichoderma sp. and tape yeast’s maximum biomass weight has a lower value than the others. This could be due to the competition that occurs between the microorganisms. Trichoderma is classified as a microorganism with high competitiveness characteristic and biofungicide activity [49]. However, this phenomenon needs to be investigated further by analyzing secondary metabolites produced during the fermentation process to obtain more conclusive explanation.
3.4. Mathematical Modelling for the Synthesis of Cellulase
In this study, mathematic modeling was carried out using secondary data from Castillo et al. [50] and Ma et al. [15] to estimate the value of kinetic parameters for production of enzyme using SMF and SSF. The value of kinetic parameters will differ from one method to another depending on microorganism species, fermentation conditions, and substrate characteristic. The secondary data from Castillo et al. [50] used a mixed culture of Trichoderma reesei and Aspergillus niger, with sorghum as the substrate, Mandel and Reese’s medium as a basal medium, and 10:1 carbon to nitrogen ratio. Fermentation process was performed at room temperature (25°C), and 70% humidity. Meanwhile in the second literature, T. reesei was cultivated in fed-batch fermentation system with 5 days of cultivation using pure cellulose as a substrate as reported b. Fermentation process was performed at 26°C, pH 5 and 30% dissolved oxygen [15].
The simulated parameters from both fermentation methods are shown in Table 3. From the results, there were differences among the parameters for SMF and SSF method. The differences could be due to the characteristic of the substrate, microorganism species, and fermentation conditions [51]. The μm value of SSF and SMF method differed from each other, although the same species of Trichoderma sp. was used in both methods. This might be related to the type of strain, fermentation method and substrate used that differ from both literatures [15]. Substrate saturation constant (Ks) of both methods also different from each other. This constant determines the saturation point of substrate at fermentation process. Both literatures use different substrates for their fermentation process. The first literature used sorghum as substrate for SSF method, meanwhile the second literature used pure cellulose as substrate for SMF method. The difference of substrate leads to different values of Ks for both simulations. Pure cellulose is well known as the best substrate for cellulase production through the fermentation process. Pure cellulose is more accessible by the microorganism to produce cellulase than another substrate such as sorghum or any agricultural waste that has another substance aside from cellulose such as lignin that could inhibit the cellulase production process by the microorganism [52].
 | Table 3: Estimated value of kinetic parameters for SSF and SMF method. [Click here to view] |
 | Figure 2: Fungal biomass weight for 5 days of fermentation at room temperature [Click here to view] |
The ccoefficient of substrate inhibition represents the concentration limit of substrate for inhibition to occur. Coefficient value is influenced by several factors such as type of substrate used, production of byproduct, and main product accumulation. Production of byproduct and accumulation of main product could inhibit enzyme production process and cell growth [53]. Ki value for SSF method was higher than SMF method. It could be influenced by the fermentation method. In solid-state fermentation, the enzyme produced by the microorganism tends to remain to stay at the substrate, whereas in SMF, the enzyme produced will be dissolved in the medium. This would lead to the higher substrate and product inhibition for the SSF method than the SMF method, resulting in a higher value of Ki at the SSF method [54].
Cell death constant, rate constant of cellulase synthesis, stoichiometric yield coefficient of biomass, and specific maintenance coefficient generally were influenced by species of microorganism used. Every microorganism has its own characteristic and metabolism pathway that could affect the value of kinetic parameters. Cell death constant describes the amount of degraded biomass. From the results presented in Table 3, a higher value of cell death constant would lead to higher value of cellulase degradation. It might be related to decreased amount of microorganism that could produce cellulase [55].
The kinetic parameters obtained from the simulation process could be used to build up mathematical model of cellulase production by SMF and SSF methods. The mathematical model consists of cell growth, substrate consumption and enzyme activity model. The simulated model compared to the experimental data to analyze the accuracy rate of the models. Comparison between simulated model and experimental data from literature for biomass, substrate, and cellulase activity was shown at Figures 3-5.
Biomass, substrate, and enzyme activity model fit the experimental data well with R2 values of 0.97; 0.97, and 0.78, respectively, for SSF method. For SMF method, R2 values for biomass, substrate, and enzyme activity model were 0.83, 0.99, and 0.93, respectively. With the high value of R2, it can be concluded that the models constructed could represent data very well and have a high accuracy rate [56]. However, further studies are needed to assess the fermentation process better with include more aspects such as mass and energy transfer that occur within the fermentation process.
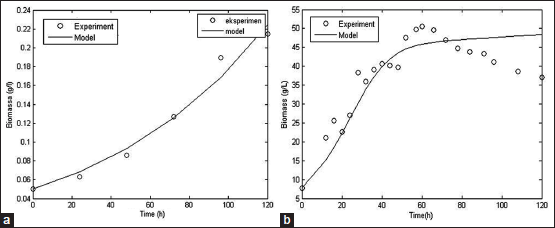 | Figure 3: Simulated model of fungal growth based on secondary data with SSF [50] (a) and SMF [15] (b) method [Click here to view] |
 | Figure 4: Simulated model of substrate consumption based on secondary data with SSF [50] (a) and SMF [15] (b) method [Click here to view] |
 | Figure 5: Simulated model of enzyme activity based on secondary data with SSF [50] (a) and SMF [15] (b) method [Click here to view] |
4. CONCLUSION
Valorization of groundnut shell as a substrate for SSF using mixed culture of Trichoderma sp., tape yeast, and tempeh yeast to produce cellulase has been studied. Lignocellulose composition of the groundnut shell has been determined and comprises 12.24% cellulose, 50.15% cellulose, and 33% lignin on a dry basis. After 5 days of fermentation, fungal biomass and cellulase activity lie in the range of 0.07–0.34 g and 0.06–0.12 FPU/ml, respectively with a maximum biomass of 0.34 g was obtained from a mixed culture of Trichoderma sp. and tempeh yeast whereas a maximum cellulase activity of 0.12 FPU/ml from a mixed culture of Trichoderma sp. and tape yeast. The results demonstrate that cellulase activity produced by mixed cultures was higher than produced by single cultures. Mathematical models were also developed using secondary data to estimate kinetic parameters for producing cellulase using submerged and SSF. Both models can predict the kinetic parameters reasonably well.
5. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
6. ACKNOWLEDGMENT
The authors would also like to thank PPMI KK ATB-ITB for the financial support provided to this study.
7. FUNDING
Research grant from Institut Teknologi Bandung (Grant number: PPMI-KK-ATB-2021).
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
7. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
8. DATA AVAILABILITY
Additional data is available upon request.
9. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Davis JP, Dean LL. Peanut composition, flavor and nutrition. In: Stalker HT, Wilson RF, editors. Peanuts Genetics, Processing, and Utilization, London: AOCS Press; 2016. p. 289-345. CrossRef
2. Naidu GK, Motagi BN, Gowda MV. Potential sources of resistance to multiple biotic stresses in groundnut (Arachis hypogaea l.). Ecoscan 2016;9:509-15.
3. Vyas AK, Putatunda C, Singh J, Vyas D. Cellulase production by Bacillus subtilis M1 using pretreated groundnut shell based liquid state fermentation. BIOTROPIA 2016;23:28-33. CrossRef
4. Kuhad RC, Singh A. Lignocellulose biotechnology: Future prospects. Crit Rev Biotechnol 2008;13:151-72. CrossRef
5. Sukumaran RK, Singhania RR, Pandey A. Microbial cellulases production, applications and challenges. J Sci Ind Res 2005;64:832-44.
6. Kuhad RC, Gupta R, Khasa YP. Bioethanol production from lignocellulosic biomass: An overview. In: Lal B, editor. Wealth from Waste. New Delhi: Teri Press; 2010. p. 53-106.
7. Pothiraj C, Balaji P, Eyini M. Enhanced production of cellulases by various fungal cultures in solid state fermentation of cassava waste. Afr J Biotechnol 2006;5:1882-5.
8. Chen H, Wang L. Technologies for Biochemical Conversion of Biomass. Cambridge: Academic Press; 2016. CrossRef
9. Bhargav S, Panda BP, Ali M, Javed S. Solid-state fermentation: An overview. Chem Biochem Eng Q 2008;22:49-70.
10. Chesson A. Effect of sodium hydroxide on cereal straws in relation to the enhanced degradation of structural polysaccharides by rumen microorganisms. J Sci Food Agric 1981;32:645-758. CrossRef
11. Vyas A, Vyas D. Production of fungal cellulases by solid state bioprocessing of groundnut shell waste. J Sci Ind Res 2005;64:767-70.
12. Ahamed A, Vermette P. Culture-based strategies to enhance cellulase enzyme production from Trichoderma reesei RUT-C30 in bioreactor culture conditions. Biochem Eng J 2008;40:399-407. CrossRef
13. Mrudula S, Murugammal R. Production of cellulase by Aspergillus niger under submerged and solid state fermentation using coir waste as a substrate. Braz J Microbiol 2011;42:1119-27. CrossRef
14. Ghose TK. Measurement of cellulase activities. Pure Appl Chem 1987;59:257-68. CrossRef
15. Ma L, Li C, Yang Z, Jia W, Zhang D. Chen S. Kinetic studies on batch cultivation of Trichoderma reesei and application to enhance cellulase production by fed-batch fermentation. J Biotechnol 2013;166:192-7. CrossRef
16. Kandala CVK, Butts CL, Lamb MC. Moisture content determination for in-shell peanuts with a low-cost impedance analyzer and capacitor sensor. Trans ASABE 2008;51:1377-81. CrossRef
17. Howard RL, Abotsi EL, Van Rensburg EJ, Howard S. Lignocellulose biotechnology: Issues of bioconversion and enzyme production. Afr J Biotechnol 2003;2:602-19. CrossRef
18. Irdhawati I, Andini A, Arsa M. The absorption of peanut shells activated by acid-base in absorbing phosphate ions in a bath with the bath method. J Kimia Riset 2016;1:52-7 CrossRef
19. Ganguly P, Sengupta S, Das P, Bhowal A. Synthesis of cellulose from peanut shell waste and its use in bioethanol production. In: Bioresource Utilization and Bioprocess. Singapore: Springer; 2020. p. 81-91. CrossRef
20. Wang B, Li D. Strong and optically transparent biocomposites reinforced with cellulose nanofibers isolated from peanut shell. Composites Part A 2015;79:1-7. CrossRef
21. Fujita M, Harada H. Ultrastructure and formation of wood cell wall. In: Wood and Cellulosic Chemistry, Revised, and Expanded. Boca Raton, Florida: CRC Press; 2000. p. 1-49.
22. Musule R, Alarcón-Gutiérrez E, Houbron EP, Bárcenas-Pazos GM, del Rosario Pineda-López M, et al. Chemical composition of lignocellulosic biomass in the wood of Abies religiosa across an altitudinal gradient. J Wood Sci 2016;62:537-47. CrossRef
23. Xu P, Cheng S, Han Y, Zhao D, Li H, Chen C. Natural variation of lignocellulosic components in miscanthus biomass in China. Front Chem 2020;8:1-12. CrossRef
24. Le Gall H, Philippe F, Domon JM, Gillet F, Pelloux J, Rayon C. Cell wall metabolism in response to abiotic stress. Plants 2015;4:112-66. CrossRef
25. Masuda Y, Kamisaka S, Yanagisawa H, Suzuki Y. Effect of light on growth and metabolic activities in pea seedlings I. Changes in cell wall polysaccharides during growth in the dark and in the light. Biochem Physiol Pflanzen 1981;176:23-34. CrossRef
26. Ricardi MM, González RM, Zhong S, Domínguez PG, Duffy T, Turjanski PG, et al. Genome-wide data (ChIP-seq) enabled identification of cell wall-related and aquaporin genes as targets of tomato ASR1, a drought stress-responsive transcription factor. BMC Plant Biol 2014;14:1-14. CrossRef
27. Piro G, Leucci MR, Waldron K, Dalessandro G. Exposure to water stress causes changes in the biosynthesis of cell wall polysaccharides in roots of wheat cultivars varying in drought tolerance. Plant Sci 2003;165:559-69. CrossRef
28. Bray EA. Genes commonly regulated by water-deficit stress in Arabidopsis thaliana. J Exp Bot 2004;55:2331-41. CrossRef
29. Kubacka-Ze?balska M, Kacperska. A. Low temperature affects pattern of leaf growth and structure of cell walls in winter oilseed rape (Brassica napus L. var. oleifera L.). Ann Bot 1999;84:313-9. CrossRef
30. Hijazi M, Velasquez SM, Jamet E, Estevez JM, Albenne C. An update on post-translational modifications of hydroxyproline-rich glycoproteins: Toward a model highlighting their contribution to plant cell wall architecture. Front Plant Sci 2014;5:395. CrossRef
31. Gutierrez-Correa M, Portal L, Moreno P, Tengerdy RP. Mixed culture solid substrate fermentation of Trichoderma reesei with Aspergillus niger on sugar cane bagasse. Bioresour Technol 1999;68:173-8. CrossRef
32. Abu Bakar NK, Abd-Aziz S, Hassan MA, Ghazali FM. Isolation and selection of appropriate cellulolytic mixed microbial cultures for cellulases production from oil palm empty fruit bunch. Biotechnology 2010;9:73-8. CrossRef
33. Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol 2010;8:15-25. CrossRef
34. Toscano PL, Correa LC, Amado MM, Ogden KL, Gómez PF. Production and Characterization of Extracellular Cellulases from a Native Strain of Trichoderma harzianum Using Wheat Straw from the Mexicali Valley as the Carbon Source. Tucson: University of Arizona; 2020.
35. Mojsov K. Application of solid-state fermentation for cellulase enzyme production using Trichoderma viride. Perspect Innov Econ Bus 2010;5:108-10. CrossRef
36. Afriyanti S, Setyabudi S. Microbial Conversion of Cassava Stem (Mannihot esculenta) Cellulose into Reducing Sugar by Trichoderma reesei PK1J2. Proceeding International Congress “Challenges of Biotechnological Research in Food and Health; 2015.
37. Scholl AL, Menegol D, Pitarelo AP, Fontana RC, Zandoná FA, Ramos L, et al. Elephant grass (Pennisetum purpureum Schum.) pretreated via steam explosion as a carbon source for cellulases and xylanases in submerged cultivation. Ind Crops Prod 2015;70:280-91. CrossRef
38. Xiang J, Wang X, Sang T. Cellulase production from Trichoderma reesei RUT C30 induced by continuous feeding of steam-exploded Miscanthus lutarioriparius. Ind Crops Prod 2021;160:113-29. CrossRef
39. Leite P, Salgado JM, Venâncio A, Domínguez JM, Belo I. Ultrasounds pretreatment of olive pomace to improve xylanase and cellulase production by solid-state fermentation. Bioresour Technol 2016;214:737-46. CrossRef
40. Goyal M, Soni G. Induction and optimization of cellulases using various agro-wastes by Trichoderma virdii: Effect of alkali pretreatment. Afr J Biotechnol 2014;13:3426-32. CrossRef
41. Singh A, Kuhad RC, Ward OP. Industrial application of microbial cellulases. In: Kuhad RC, Singh A, editors. Lignocellulose Biotechnologgy: Future Prospects. New Delhi, India: I. K. International Publishing House; 2007. p. 345-58.
42. Rahikainen JL, Martin SR, Heikkinen H, Rovio S, Marjamaa K, Tamminen T, et al. Inhibitory effect of lignin during cellulose bioconversion: The effect of lignin chemistry on non-productive enzyme adsorption. Bioresourc Technol 2013;133:270-8. CrossRef
43. Zhao CH, Liu X, Zhan T, He J. Production of cellulase by Trichoderma reesei from pretreated straw and furfural residues. RSC Adv 2018;8:36233-8. CrossRef
44. Amriani F, Salim FA, Iskandinata I, Khumsupan D, Barta Z. Physical and biophysical pretreatment of water hyacinth biomass for cellulase enzyme production. Chem Biochem Eng Q 2016;30:237-44. CrossRef
45. Nisizawa T, Suzuki H, Nakayama M, Nisizawa K. Inductive formation of cellulase by sophorose in Trichoderma viride. J Biochem 1971;70:375-85. CrossRef
46. Jourdier E, Cohen C, Poughon L, Larroche C, Monot F, Chaabane FB. Cellulase activity mapping of Trichoderma reesei cultivated in sugar mixtures under fed-batch conditions. Biotechnol Biofuels 2013;6:1-12. CrossRef
47. Aro N, Ilmén M, Saloheimo A, Penttila? M. ACEI of Trichoderma reesei is a repressor of cellulase and xylanase expression. Appl Environ Microbiol 2003;69:56-65. CrossRef
48. Lin D, Lopez-Sanchez P, Li R, Li Z. Production of bacterial cellulose by Gluconacetobacter hansenii CGMCC 3917 using only waste beer yeast as nutrient source. Bioresour Technol 2014;151:113-9. CrossRef
49. Kolombet LV, Zhigletsova SK, Kosareva NI, Bystrova EV, Derbyshev VV, Krasnova SP, et al. Development of an extended shelf-life, liquid formulation of the biofungicide Trichoderma asperellum. World J Microbiol Biotechnol 2008;24:123-31. CrossRef
50. Castillo MR, Gutierrez-Correa M, Linden JC, Tengerdy RP. Mixed culture solid substrate fermentation for cellulolytic enzyme production. Biotechnol Lett 1994;16:967-72. CrossRef
51. Borchers S, Freund S, Rath A, Streif S, Reichl U, Findeisen R. Identification of growth phases and influencing factors in cultivations with AGE1. HN cells using set-based methods. PLoS One 2013;8:1-11. CrossRef
52. Agbor VB, Cicek N, Sparling R, Berlin A, Levin DB. Biomass pretreatment: Fundamentals toward application. Biotechnol Adv 2011;29:675-85. CrossRef
53. Srinivasan K, Murakami M, Nakashimada Y, Nishio N. Efficient production of cellulolytic and xylanolytic enzymes by the rumen anaerobic fungus, Neocallimastix frontalis, in a repeated batch culture. J Biosci Bioeng 2001;91:153-8. CrossRef
54. Hölker U, Höfer M, Lenz J. Biotechnological advantages of laboratory-scale solid-state fermentation with fungi. Appl Microbiol Biotechnol 2004;64:175-86. CrossRef
55. Dey P, Pal P. Modelling and simulation of continuous L (+) lactic acid production from sugarcane juice in membrane integrated hybrid-reactor system. Biochem Eng J 2013;79:15-24. CrossRef
56. Onyutha C. From R-squared to coefficient of model accuracy for assessing goodness-of fits. Geosci Model Dev Discuss 2020;511-25. CrossRef