1. INTRODUCTION
The transcriptional control of various genes required for developing and maintaining bacteria-host relationships depends on bacterial quorum sensing (QS) [1]. It is the capacity of gene regulation to recognize and react to cell population density. QS, for instance, allows bacteria to control the expression of particular genes to the high densities of cells where the associated phenotypes will be most advantageous [2]. Bacteria use acyl-homoserine lactones (Gram-negative bacteria), peptides (Gram-positive bacteria), and Furanosyl borate diester as QS signaling molecules. These molecules are called as autoinducers [3].
Pseudomonas aeruginosa is a Gram-negative, rod-shaped, encapsulated bacterium that causes opportunistic infections and frequently infects immunocompromised individuals who are already suffering from serious diseases, most notably cystic fibrosis and severe burns [4]. The regulation of pathogenicity, virulence, and development of biofilm is greatly influenced by QS [5]. There are mainly four types of QS systems found in P. aeruginosa, las, rhl, Pseudomonas quinolone signal (PQS), and integrated QS which together form an interconnected network [6,7]. Out of these four QS systems, the Las system was found to be the most significant in controlling the QS system. LasI and LasR are the two main components of the Las system. LasI is responsible for producing 3-oxo-C12-HSL which acts as an autoinducer molecule. 3-oxo-C12-HSL binds to LasR and triggers the transcriptional control of numerous genes [7,8]. LasR is the main protein that controls various virulence factors, such as the formation of biofilm and the production of virulence factors such as pyocyanin, proteases, exotoxins, and rhamnolipids [9–11]. Structurally, at the C terminal of LasR, a DNA binding domain is found, whereas, a ligand binding domain is present at the amino-terminal [12,13]. When 3-oxo-C12-HSL binds to the LasR, its monomeric form dimerizes to form two LasR subunits after the stabilization of LasR by 3-oxo-C12-HSL. As a result, this dimeric form of LasR binds to DNA and activates the cascade of virulence factors [14,15]. Therefore, LasR plays a crucial role in QS associated pathogenicity of P. aeruginosa.
Over the years, several methods have been tried to find a potent inhibitor having anti-QS and anti-biofilm activity against P. aeruginosa. Initially, the biosensor was used to identify the potential Quorum Sensing Inhibitor (QSI), and these biosensors have QS-regulated promotors linked to a receptor gene such as lacZ, lux, and gfp [16–18]. Although various QSI were identified by biosensor systems, there is the possibility that they may target other regulators that can positively impact the QS system and, thereby, activate the virulence factors [19]. Another approach is a computer-based method for the identification of potential QSI. It can be more precise and reduce the risk associated with biosensor systems [19]. The compounds identified by in silico methodologies can be checked for their efficacies by molecular docking and dynamic simulation [20]. As a result, the chances of rejection of these compounds are relatively reduced in clinical trials.
In this study, we have screened an entire library of phytochemical compounds obtained from the Zinc database for their anti-QS potential against P. aeruginosa. The transcriptional regulator LasR was selected as a target due to its vital role in the QS system. The pharmacokinetic properties of the QSIs with substantially higher docking scores were chosen and examined.
2. MATERIALS AND METHODS
2.1. Preparation of Protein
The 3D structure of LasR, having a resolution of 1.4 A was obtained from the Protein Data base (ID 3ix3). The 3D structure obtained from Protein Data Bank cannot be used for docking because it contains native ligands, heteroatoms, and water molecules. Hence before performing docking analysis, heteroatoms and their native ligand, along with water molecules are removed, and hydrogen is added.
2.2. Preparation of Ligand
A phytochemical library of 2,068 natural compounds was selected from the ZINC library, and we have ensured that these compounds are commercially available [21]. The structural coordinates of the compounds were obtained from the database in .Sdf format [22]. Now open Babel software was used to convert these compounds from 2D to 3D form.
2.3. Virtual Screening
We have used PyRx software for virtual screening [23] because it contains both autodock [24] and autodock vina [25]. PyRX scoring system used the Lamarckian genetic method to dock different compounds to LasR. In this present study, we have used Autodock for molecular docking. The grid box was formed using dimensions X: 33.87, Y: 46.69, and Z: 54.92 A0 with active side residue. To simulate biological conditions, docking was carried out in the presence of neutralizing ions and water molecules. After docking, binding energy was used to rank all the molecules.
2.4. ADME and Toxicity Studies
To determine drug safety, an ADMET study was performed using SWISS ADME. Before this analysis, the ligands were selected based on their molecular weight, number of hydrogen bond donors, and Lipinski rule of five [26–29]. With the help of Swiss ADME, the molecules were analyzed for their various pharmaceutical properties, such as GI adsorption and BBB permeability. After this Lazar toxicity module was applied for further screening of molecules for their toxicity properties [30].
 | Table 1. Selected molecules with their respective ZINC ID and ADME properties. [Click here to view] |
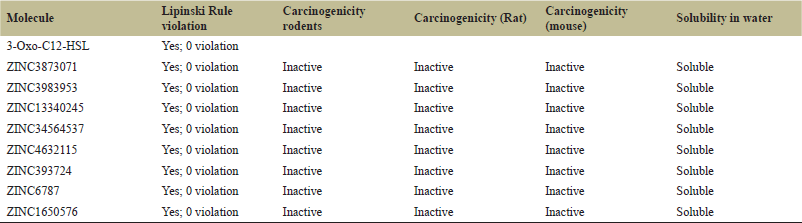 | Table 1 continues [Click here to view] |
 | Table 2. Screened molecules with their binding energy, number of hydrogen bonds formed and residue involved in hydrogen bonding. [Click here to view] |
2.5. Molecular Docking
Only those ligands were picked for docking studies against active sites of LasR that have passed the ADME and toxicity filters to determine the optical inhibitor among all the screened molecules. For this, we have used Autodock4 software. Again, the grid box was used to define the protein’s active site. Lamarckian genetic algorithm was used to dock the into the receptor’s grid. Binding energy was calculated by the following equation:
ΔG= (VboundL-L – VunboundL-L) + (VboundP-P –VunboundP-P) + (VboundP-L – VunboundP-L +ΔSconf)
Here, P = protein, L = ligand, V = pairwise evaluations, and ΔSconf = loss of conformational entropy upon binding.
3. RESULT AND DISCUSSION
In this study, four different filters were utilized to identify the most efficient QSI. First, we have PyRx as the primary filter. After this, we selected only those molecules for secondary screening with binding energy lesser than −8. Molecules having lesser energy than −8 were screened by Lipinski’s rule of five, which acts as our secondary filter. Then, the molecules were selected based on the H bond donor and acceptor, and finally, all the molecules were screened based on their ADME and toxicity analysis.
3.1. Virtual Screening
PyRx was used for the secondary screening of molecules obtained from the Zinc library. A total of 599 molecules were taken for further study as they have binding energy lesser than −8. The native ligand of LasR binds to the active site of its receptor protein with a binding energy of −6.5, so we selected only those molecules with binding energy less than −8. Hence, they show effective binding with LasR.
3.2. ADME and Toxicity Filters
457 compounds passed from Lipinski’s rule of five, but only 274 compounds were further selected based on their number of hydrogen bonds. Molecules with unfavorable hydrogen bonds were not taken for further consideration. After this, Swiss ADME software was used for the ADME studies, molecules were sorted based on their gastrointestinal absorption, blood–brain barrier permeability, and skin permeation. A total of 53 molecules were selected based on the ADME filter. Now, all the molecules were subjected to toxicity studies, and only eight were found safe in their toxicity profile. Table 1 shows all the molecules with their ZINC ID and other properties.
To understand the molecular interaction, we have again docked these molecules against the active site of LasR.
3.3. Molecular Docking
ZINC3873071, ZINC3983953, ZINC13340245, ZINC34564537, ZINC4632115, ZINC393724 ZINC6787, and ZINC1650576 and were selected for the final study. Native ligand of LasR, i.e., 3-oxo-C12HSL was also docked against its active site. The binding energy of 3-oxo-C12-HSL to LasR was reported to be −6.5. Table 2 shows the selected molecules with their binding energy, number of hydrogen bonds, and residue involved in binding.
3.4. Analysis of Interactions
Figure 1A and B shows the binding of ZINC3873071 to protein 3ix3. Here, protein-ligand complex is stabilized by three interactions that involve hydrogen bonds, alkyl, and Van der Waals interaction with a binding energy of −10.3. Residue Trp60, Trp88, and Ser129 are involved in hydrogen bonding. The interaction between 3ix3 and ZINC3983953 is stabilized by the formation of three hydrogen bonds between the ligand and Arg61, Asp65, and Thr75 of the receptor molecule with a binding energy of −9.3 (Fig. 2). ZINC13340245 interacts with protein 3ix3 with three strong hydrogen bonds. The residues involved in the hydrogen bond formation are Asp65, Thr75, and Thr115 with a binding energy of −9.5 (Fig. 3A and B). Figure 4A and B shows the interaction of ZINC34564537 to protein 3ix3. The binding energy of this interaction was found to be −8.6 and two hydrogen bonds stabilized the interaction. ZINC4632115 was docked against protein LasR with a binding energy of −8.4. This binding was stabilized by three hydrogen bonds and alkyl interactions (Fig. 5A and B). Protein 3ix3 and ZINC393724 molecules interact with each other, and the binding energy of this interaction was found to be −8.4 (Fig. 6).
Two hydrogen bonds formed between ZINC6787 and protein 3ix3 for its stabilization. Figure 7A and B shows that the Tyr93 and Leu110 of receptor protein are involved in hydrogen bonding with a binding energy of −9.1. ZINC1650576 and protein were stabilized by two hydrogen bonds with a binding energy of −10.7, and Trp88 and Ser129 residues are involved in the hydrogen bond formation between ligand and receptor (Fig. 8). Figure 9A and B shows the interaction between LasR and its native ligand 3-oxo-C12-HSL. The binding energy of this interaction was found to be −7.5 and the interaction was stabilized by four hydrogen bonds formed between Tyr56, Trp60, Asp73, and Ser129 with the ligand.
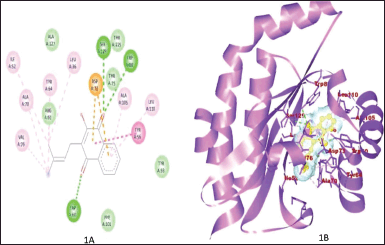 | Figure 1. A and B show the 2D and 3D interaction between molecule ZINC3873071 and protein 3ix3, respectively. [Click here to view] |
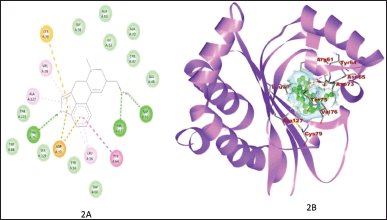 | Figure 2. 2D (A) and 3D (B) splots demonstrating the interaction of ligand ZINC3983953 with the active site of receptor protein (3ix3) and interaction was stabilized by three H-Bonds. [Click here to view] |
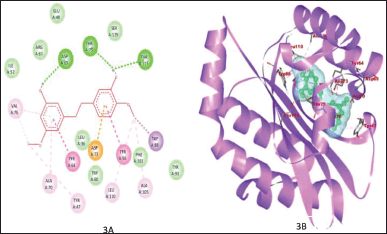 | Figure 3. 2D plot showing the interaction of ligand ZINC13340245 and 3ix3 (A) and 3D plot demonstrating that Asp65, Thr75, and Thr115 are involved in Hydrogen bonding (B). The binding was also stabilized by alkyl and van der Waals interaction. [Click here to view] |
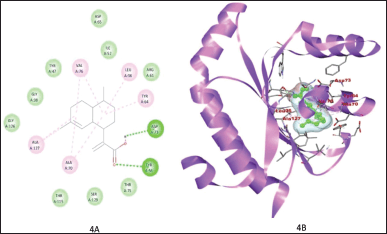 | Figure 4. 2D and 3D plot depicting the interaction between ligand ZINC34564537 and its receptor. This binding was stabilized by two strong H bond, alkyl interaction and van der Waals interaction. Asp73 and Tyr56 residue of receptor protein involved in the hydrogen bonding with ligand. [Click here to view] |
 | Figure 5. (A) shows the 2D plot demonstrating the interaction of ligand ZINC4632115 and receptor molecule while (B) shows the 3D plot of the same interaction. Here, three residues of the receptor molecule, i.e., Trp60, Trp88, and Ser129 are involved in the hydrogen bonding with the ligand molecule. [Click here to view] |
The native ligand 3 oxo binds with the lasR with four hydrogen bonds. These hydrogen bonds are formed between Ser129, Thr56, Asp73, and Trp60 at the receptor’s active site. We also found that Tyr and Ser are the two most common residues of the receptor, which are involved with almost all of the docked ligands. When we compared the binding energy of all the docked molecules, we found that it was better than its native ligand 3-oxo-C12-HSL. We have found that the molecular weight of ZINC 18847036 (268.26), ZINC13340245 (274.31), and ZINC 1650576 (294.34) was almost similar to that of 3-oxo-C12-HSL (268.5). Here, in this study, we report that all these molecules have different structures from that of native signaling molecules; hence, they can be a promising candidate for the synthesis of novel drugs. LasR inhibitors have been discovered by several groups using computer-based drug designing. Due to this, it is possible to identify novel LasR inhibitors that can reduce the virulence pathogenicity and resistance of the infection without significantly affecting the growth kinetics of bacteria.
Xu et al. [31] screened 2,030 natural compounds against coronavirus (COVID-19) based on molecular docking and then studied the six closest analogues for their activity against coronavirus. In 2021 Vetrivel et al. [6] performed high throughput virtual screening of Schrodinger small molecules database. They took 12 best hits, and out of these 12, they found three molecules that may serve as quorum-sensing inhibitors against P. aeruginosa. Annapoorani et al. [32] also screened 1,920 natural compounds against P. aeruginosa; four compounds were found to be very effective against it. Baicalein (Supplementry figure1) a known inhibitor of QS in P. aeruginosa, is also a natural compound that was discovered by Zeng et al. [33].
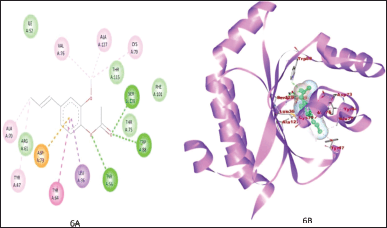 | Figure 6. 2D (A) and 3D (B) plot showing the interaction between ligand ZINC393724 and active site of protein. The binding was stabilized by three Hydrogen bond and residue involved are Trp60, Trp88, and Ser129. [Click here to view] |
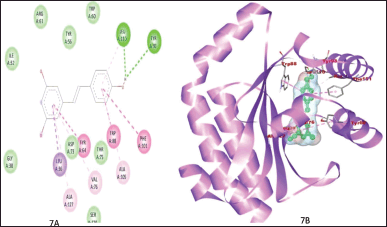 | Figure 7. 2D and 3D plot showing the interaction between ZINC6787 and protein 3ix3. Besides two hydrogen bonds this interaction was also stabilized by alkyl and van der Waals interactions. [Click here to view] |
 | Figure 8. (A) shows the 2D plot while (B) shows the 3D plot of interaction between ligand ZINC1650576 and protein. The binding was stabilized by two strong hydrogen bonds. [Click here to view] |
 | Figure 9. 2D and 3D plot demonstrating the binding pattern of the native ligand of LasR with the protein 3ix3. Here, total four hydrogen were formed between ligand and protein to stabilize the interaction. The green dot represents the hydrogen bonding. [Click here to view] |
 | Supplementary Figure 1. 2D and 3D plot demonstrating the binding pattern of the Baicalein with the protein 3ix3. Here, total four hydrogen were formed between ligand and protein to stabilize the interaction. The green dot represents the hydrogen bonding. [Click here to view] |
4. CONCLUSION
Prevention of bacterial infection is severely hampered by biofilm formation, and the situation becomes even more adverse when the patient is suffering from Urinary tract infection, cystic fibrosis, or burn injury. Pseudomonas aeruginosa is an opportunistic pathogen that is a leading cause of hospital-acquired infections. In the pathogenesis of P. aeruginosa, LasR plays a very significant role as a transcription factor as it controls the cascade of virulence genes. One of the significant attributes of P. aeruginosa is biofilm formation (governed by lasR) in the disease condition. Due to biofilm formation, there is an enhanced multidrug resistance to antibiotics because the membrane permeability of the antibiotics is significantly reduced. The current investigation aimed to find the new QS inhibitors. To accomplish this task, we have screened a phytochemical library containing 599 compounds from the ZINC database against the master regulator of QS, which is LasR. After docking, we were able to identify eight potential molecules that show anti-QS activity against P. aeruginosa. These molecules were further screened for their drug-like properties, which revealed their drug likeness. Being of natural origin, these molecules open the door for developing possibly safe and affordable QSI as therapeutics. The screened compounds need to be validated through in vitro and in vivo studies following the computational methods.