1. INTRODUCTION
The Lamiaceae is a large family composed of 264 genera and around 7000 described species [1]. Square-like stems, whorl- or oppositely-positioned leaves, and irregular or zygomorphic flowers, usually with a 2-lipped corolla, characterize plants belonging to this family. Species of this family are mostly herbs or shrubs and rarely trees. They are endemic in two main centers of biodiversity: the Mediterranean basin, central Asia, and subtropical regions [2]. The members of this family are known as medicinal and aromatic herbs such as mint, sage, thyme, basil, rosemary, lavender, and oregano, which have been widely used as spices, teas, or traditional medicines [3-6]. In Saudi Arabia and the Arab peninsula, some of these species, including Mentha are commonly used as hot drinks or tea, home-based ailment relievers, or spices for cooking due to their contents of aromatic oils and other secondary metabolites [7]. Therefore, many species of the Lamiaceae family are considered commercial commodities, such as the Madinah mint in Saudi Arabia. Many species and cultivars of this family have been investigated for biochemical properties or taxonomic purposes [8-14]. While specific research on Madinah mint is limited, studies suggest that it possesses antioxidant, antimutagenic, and anticancer properties [15,16], and studies on related mint species suggest that it may possess antioxidant, antibacterial, antifungal, antiviral, and anticancer properties [17]. Further research is needed to fully understand the unique properties and potential health benefits of Madinah mint.
DNA barcoding is a technique developed by Hebert et al. [18] that relies on universal primers to amplify and sequence short universal DNA sequences. This technique has recently been applied as a universal tool for species authentication and identification [19-22]. The basic idea of this technique is to generate a standardized short DNA sequence(s) from any small tissue sample of a plant and compare it to any institutional library or international databases that contain reference sequences of the same or related species. This process will provide a rapid and reproducible taxonomic identification [18,23-29]. This method has been applied to identify members within the family Lamiaceae as well as to identify commercial processed spice species belonging to this family [10,11,30,31]. Herbs and spices are among trade commodities that can be adulterated, intentionally or accidentally, with morphologically similar plants. Hence, DNA barcoding would be a suitable technique to investigate and characterize these commercialized herbs possibly to the genus or species level [32-38].
DNA barcoding in animals is well established [39-41], but in plants, it is important to search for a suitable genomic region to perform DNA barcoding. However, some regions were suggested by scientists from around the Globe as well as the Plant Working Group of the Consortium for the Barcode of Life [42-44]. The suggested DNA regions include coding sequences from the plastid genome such as matK, rbcL, rpoB, rpoC1, and ycf5, genes as well as intergenic spacers such as trnH-psbA, atpF-atpH, and psbK-psbI. Moreover, the nuclear ITS1 and ITS2 have also been proposed as efficient plant DNA barcoding regions [45-51]. Recently, combinations of matK + rbcL or ITSs were proposed by the CBOL Plant Working Group (https://ibol.org) to increase the efficiency of plant species identification [43,52]. Even though a combination of the most appropriate regions for plant DNA barcoding and identification remains contentious and relies on trial and error [26,53-56].
The objective of this study is to investigate the applicability of DNA barcoding on the locally traded and available Madinah mint for authentication and identification. This study provided an evaluation of seven single candidate DNA barcoding loci and some of their combinations to identify the traded Madinah mint in Saudi Arabia.
2. MATERIALS AND METHODS
2.1. Plant Samples
Samples from traded Madinah mint were bought from the local market (Al-Ahsa, Saudi Arabia), which were used to test the performance of seven different candidate genomic regions for DNA barcoding analyses [Table 1].
Table 1: Primers and PCR conditions for the seven selected DNA barcoding loci tested against Madinah mint in the present study [53].
| Locus | Primer’s Name | Primer sequences (5’- 3’) | Length (bp) | PCR cycle conditions | Cycles |
|---|---|---|---|---|---|
| matK | matKF | CgATCTATTCATTCAATATTTC | 23 | 95°C 1 min | 35 |
| matKR | TCTAgCACACgAAAgTCgAAgT | 22 | 50°C 30 s | ||
| 72°C 1 min | |||||
| rbcL | rbcLF | ATgTCACCACAAACAgAAAC | 20 | 95°C 1 min | 35 |
| rbcLR | TCgCATgTACCTgCAgTAgC | 20 | 55°C 30 s | ||
| 72°C 1 min | |||||
| trnA-H | trnA-HF | gTTATgCATgAACgTAATgCTC | 22 | 95°C 1 min | 35 |
| trnA-HR | CgCGCATggTggATTCACAATCC | 23 | 55°C 30 s | ||
| 72°C 1.5 min | |||||
| rpoC1 | rpoCF | ggCAAAgAgggAAgATTTCg | 20 | 95°C 1 min | 40 |
| rpoCR | CCATAAgCATATCTTgAgTTgg | 22 | 53°C 40 s | ||
| 72°C 40 s | |||||
| ycf5 | ycf5F | ACTTTAgAgCATATATTAACTC | 22 | 95°C 1 min | 40 |
| ycf5R | ACTTACgTgCATCATTAACCA | 21 | 53°C 40 s | ||
| 72°C 40 s | |||||
| ITS1 | ITS1F | CCTTATCATTTAgAggAAggAg | 22 | 95°C 1 min | 35 |
| ITS1R | TCCTCCgCTTATTgATATgC | 20 | 50°C 30 s | ||
| 72°C 1.5 min | |||||
| ITS2 | ITS2F | ATgCgATACTTggTgTgAAT | 20 | 95°C 1 min | 40 |
| ITS2R | gACgCTTCTCCAgACTACAAT | 21 | 56°C 30 s | ||
| 72°C 45 s |
PCR: Polymerase chain reaction
2.2. DNA Isolation and Amplification
A sterile mortar and pestle were used to crush dry leaves (100 mg) under liquid nitrogen for DNA extraction. Dry leaves were used instead of fresh ones due to availability; they are more accessible in the Al-Ahsa region in eastern Saudi Arabia, whereas Madinah is located in the west. DNA was isolated using the Plant Genomic DNA Extraction Miniprep System (Viogene BioTek Corp., Taipei, Taiwan) to obtain high-quality DNA, free of polysaccharides or other metabolites that might interfere with DNA amplification. The protocol of the manufacturer was followed. Purified DNA concentration of samples was estimated both fluorometrically using a NanoDrop 2000c instrument (Thermo Scientific, DE, USA) and by comparison of ethidium bromide-stained band intensities with DNA standard (Edvotek, Washington, USA). The extracted DNA purity was above 1.83 at A260/A280.
2.3. Polymerase Chain Reaction (PCR) Amplification
PCR amplification for each candidate locus was performed using GoTaq® DNA Polymerase (Promega, CA, USA) in a 25 μL reaction volume according to the manufacturer’s instructions. PCR protocols for the seven selected loci are listed in Table 1 that are all started with a denaturation step of 2 min at 95°C and ended with a final extension step at 72°C for 7 min.
One percent agarose gel size 7 × 7 cm was prepared for electrophoreses of amplified PCR products in ×1 TAE buffer. The gel was stained with ethidium bromide (0.5 μg/mL) in ×1 TAE buffer. Gel images were obtained using Benchtop 3UVTM transilluminator equipped with a BioDoc-It Imaging System (UVP, CA, USA). The size and presence or absence of amplified PCR products were determined on gel using a standard DNA ladder (Edvotek, Washington, USA). The ladder shortest fragment is 570 bp, and the longest is 23130 bp.
2.4. DNA Sequencing
PCR-amplified DNA was purified and bi-directionally sequenced by Macrogen Inc., Korea (http://www.macrogen.com). Forward and reverse sequences were obtained using the same primers that were used for PCR amplification, manually edited, and the 3’ and 5’ terminals were clipped to generate consensus sequences for each locus.
2.5. Data Analysis and Madinah Mint Delimitation
The Basic Local Alignment Search Tool (BLAST) [57] from the National Center of Biotechnology Information (NCBI) was used to search for relevant sequences in the NCBI and Barcode of Life Data databases. Clustal W [58] and Genetyx software (Genetyx, Tokyo, Japan) were used to align sequences obtained from the tested species and relevant sequences that were retrieved from the international databases. Pair-wise sequence comparisons of closely related plant species were conducted using BLAST2 Sequences [59] and Genetyx software (Genetyx, Tokyo, Japan). The Neighbor-Joining method of MEGA 11 [60] was used for phylogenetic analyses. The topologies of the phylogenetic trees were evaluated using the bootstrap re-sampling method of Felsenstein [61] and 1000 replicates. Phylogenetic trees have been built by applying the data to MEGA 11 [60], and the options were as follows; Statistical method: Neighbor-Joining, Test of phylogeny: Bootstrap method (1000 replicates), Model/Method: p-distance, Substitutions to Include: d: Transitions + Transversions, Rates among Sites: Uniform rates, Pattern among Lineages: Same (Homogeneous), Gaps/Missing Data Treatment: Pairwise deletion, and Select Codon Positions: 1st + 2nd + 3rd + non-coding.
3. RESULTS
3.1. Amplification and Sequencing of Seven DNA Barcoding Candidate Loci
A good yield of high-quality DNA was obtained from the Madinah mint samples under study. The first step of this work was to empirically test the universality of seven DNA barcoding candidate loci [Table 1]. Therefore, PCR amplification was conducted in different trials. The first trial was performed under standard PCR conditions starting from around 50 ng of template DNA. The second trial was applied only on loci that generated multiple and/or non-specific PCR products or did not generate any amplicons, where higher annealing temperature was applied. Loci and template DNA samples that failed to amplify were tried under lower stringency conditions through reduced annealing temperature and/or increased number of cycles. In case of failure of both trials, 0.5 or 1 μL of PCR products from both trials were then used as template DNA and re-amplified. PCR was considered a failure only in case of negative amplifications under all these different conditions. Samples of traded Madinah mint tested against the seven selected loci exhibited 100% PCR amplification success after applying these different trials [Figure 1]. The amplified PCR products of the seven DNA loci were successfully sequenced, and high-quality bi-directional sequences were obtained. Sequences were submitted to the NCBI database, and accession numbers were acquired (matK; PV031553, rbcL; PV031554, rpoC1; PV031555, ycf5; PV031556, ITS1; PQ836415, ITS2; PQ836431).
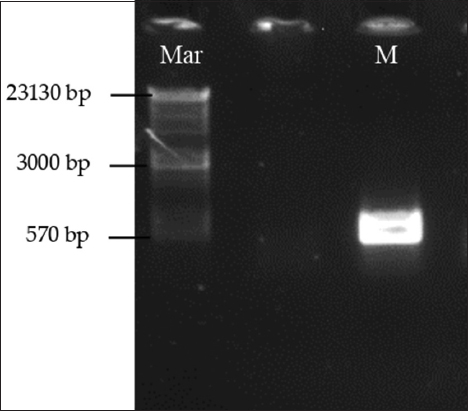 | Figure 1: Polymerase chain reaction amplification of ITS2 from Madinah mint (M). (Mar) refers to a DNA ladder (Edvotek, Washington, USA). The ladder shortest fragment is 570 bp and the longest is 23130 bp. [Click here to view] |
3.2. Data Analysis
Based on a homology search of the database using sequences obtained from the plastid matK gene, Madinah mint is 100% (766bp) identical to M. piperita, but very similar to M. spicata, Mentha aquatica, Mentha longifolia, Mentha pulegium and M. suaveolens (99.9%; 753-68 bp), some transitions/transversions were observed [Table 2]. Using the matK gene sequence from Madinah mint as a reference, a transition from (G) to (A) at position 641 was observed in M. canadensis, M. spicata, and M. suaveolens, from (A) to (G) in M. aquatica at position (5), from (C) to (T) in M. longifolia at position 430, and from (T) to (C) in M. canadensis at position 595. One transversion from (A) to (T) was observed in M. pulegium at position 21. An indel (insertion/deletion) of 6 bp direct repeat was also observed in Mentha cervina and Mentha arvensis at the same position (76–81), which indicates that these later species share the same ancestral plastid genome that is different from other species, at least in this part of DNA.
Table 2: Sequence identity, transitions, transversions, and indels of Madinah mint under study compared with other Mentha species based on sequences of matK or rbcL gene.
| Species | Base | M. canadensis | M. spicata | M. aquatica | M. longifolia | M. piperita | M. pulegium | M. suaveolens | M. cervina and M. arvensis |
|---|---|---|---|---|---|---|---|---|---|
| matK | |||||||||
| 99.7% (768 bp) | 99.9% (768 bp) | 99.9% (768 bp) | 99.9% (753 bp) | 100% (766 bp) | 99.9% (768 bp) | 99.9% (768 bp) | 95.7 and 99.1% (768 bp) | ||
| Madinah Mentha | A | G (5) | T (21) | ||||||
| G | A (641)* | A (641) | A (641) | ||||||
| C | T (430) | ||||||||
| T | C (595) | ||||||||
| Indel (TTGGAA) (76–81) | |||||||||
| rbcL | |||||||||
| 100% (670 bp) | 100% (666 bp) | 99.7% (670 bp) | 99.7% (670 bp) | 99.9% (670 bp) | |||||
| A | C (616) | G (311) | |||||||
| G | |||||||||
| C | T (81) | ||||||||
| T | G (625) | C (438) | |||||||
When homology search of the NCBI database was performed using sequences obtained from the plastid rbcL gene of Madinah mint as a query, it was 100% (666-70 nt) identical to M. canadensis and M. spicata as well very closely related to M. suaveolens (99.9%; 670 bp) with only one base transition from (T) to (C) at position 438. However, it is also related to M. longifolia (99.7%; 670bp) with two transitions from (C) to (T) at position 81 and (A) to (G) at position 311. A base identity of 99.7% (670bp) was also detected between Madinah mint and M. aquatica with two transversions of (A) to (C) at position 615 and (T) to (G) at position 625 [Table 2].
When BLAST was applied to search the NCBI database using sequences obtained from the intergenic region trnA-trnH from Madinah mint as a query, it did not retrieve any Mentha spp. similarities except for Chenopodium spp. and other related taxa with base identity ranging between 80.8% in Chenopodium foliosum and 87.2% in Chenopodium album. Sequences of the rpoC1 gene from Madinah mint retrieved a single similarity that is M. aquatica (99.7% base identity), where there is only one transversion from (C) to (A) at position 148 [Figure 2]. The ycf5 sequences of Madinah mint resulted in similarities to M. x piperita, M. spicata, and M. canadensis (98.92%) [53].
 | Figure 2: Sequences alignment of rpoC1 from Madinah mint and M. aquatica reveals a single single nucleotide polymorphism. [Click here to view] |
Homology search of the database using sequences of ITS1 distanced Madinah mint from M. pulegium and M. piperita (93.2% identity; 585-9 bp) as well as M. arvensis (93.4% identity; 580bp), but M. suaveolens scored 97.8% identity (584 bp). Yet, Madinah mint remained closely related to M. spicata and M. canadensis (98.1 and 98.5% identity, respectively; 584 bp). Likewise, a homology search of ITS2 sequences showed that Madinah mint is distant from M. pulegium and M. piperita (92.4 and 92.9% identity, respectively; 302-50 bp) as well as M. suaveolens (96.8% identity; 312 bp), but remained closely related to M. spicata and M. canadensis (99.4% identity; 340-50 bp).
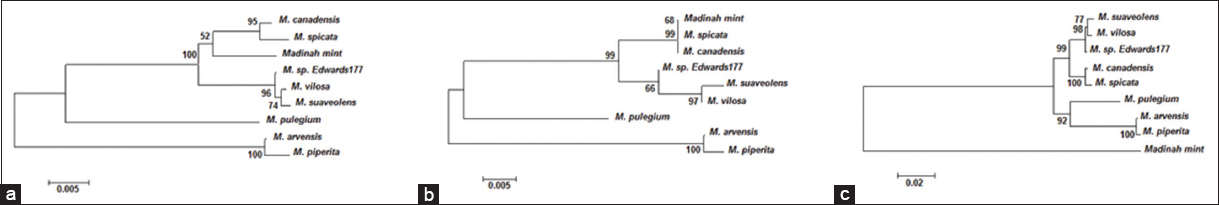 | Figure 3: Evolutionary relationships of taxa inferred using the Neighbor-Joining method [87] based on (a) matK, (b) rbcL, and (c) matK + rbcL sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [61]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method [88] and are in the units of the number of base differences per site. Codon positions included were 1st + 2nd + 3rd. All ambiguous positions were removed for each sequence pair. Evolutionary analyses were conducted in MEGA 11 [60]. [Click here to view] |
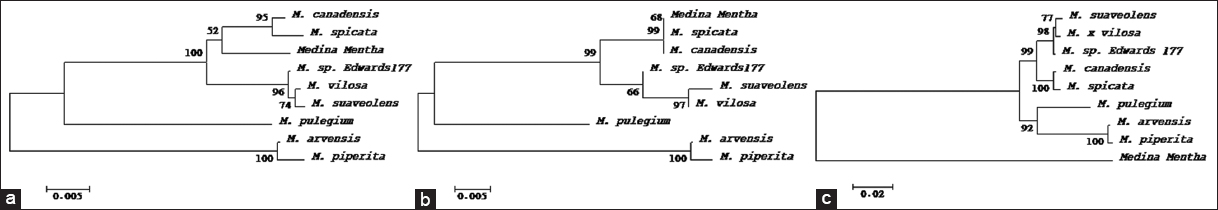 | Figure 4: Evolutionary relationships of taxa inferred using the Neighbor-Joining method [87] based on (a) ITS1, (b) ITS2, and (c) ITS1 + ITS2 sequences. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches [61]. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method [88] and are in the units of the number of base differences per site. All ambiguous positions were removed for each sequence pair. Evolutionary analyses were conducted in MEGA 11 [60]. [Click here to view] |
4. DISCUSSION
The BLAST, genetic distance and tree topology are some of the techniques applied to compare DNA sequences obtained from different living organisms for the purpose of molecular identification and delimitation [55,62-65]. DNA barcoding is a developed technique that uses specific DNA sequences to identify living organism species [18]. In plants, several DNA loci, either from the plastid organelle or nuclear genomes, were proposed by the Plant Working Group of the Consortium for the Barcode of Life [42-44]. These suggested DNA barcodes included coding sequences from matK, rbcL, rpoB, rpoC1, and ycf5 genes, as well as trnH-psbA, atpF-atpH, and psbK-psbI intergenic spacers of the plastid genome. Besides, the nuclear ITS1 and ITS2 have also been proposed as efficient plant DNA barcoding loci [43,45,47-50,66]. Another suggestion from the CBOL Plant Working Group (www.barcoding.si. edu/plant_working_group. html) is to use combinations of matK + rbcL or ITSs for DNA barcoding so as to increase the efficiency of plant species identification [43,67,68]. Yet, a combination of two or more appropriate loci for plant DNA barcoding remains debatable and requires multiple trials [26,53,54,69-71].
In this study, partial coding sequences from matK, rbcL, rpoC1, and ycf5 genes, beside noncoding sequences from the intergenic space trnA-trnH as well as ITS1 and ITS2 were tested against one of the most traded and famous Mentha in Saudi Arabia; Madinah mint. Sequences obtained from these loci of Madinah mint were applied to BLAST search on the international database.
The matK gene is often challenging to amplify and sequence using PCR due to its high sequence variability and complex secondary structure [44,72-75]. This study has successfully amplified and sequenced this region from the Madinah mint and was able to differentiate it from other Mentha species available on the database, except for M. piperita, which was 100% identical. The matK sequence of Madinah mint showed a single unique single nucleotide polymorphism (SNP) with M. spicata, M. aquatica, M. longifolia, M. pulegium, and M. suaveolens and 2 SNPs with M. canadensis [Table 2]. These SNPs could be very useful in the authentication and identification of Mentha species. SNPs were applied in authentication and identification of other plant species such as Morinda umbellata and Matelea reticulata [76], Coffea canephora and Coffea congensis [77], Diospyros mespiliformisand Diospyros brandisiana [28] and Patrinia species [78]. Phylogenetic analysis using the Neighbor-Joining method with partial sequences from matK gene was able to reveal the molecular evolution of Madinah mint that formed a basic phylogenetic clade and seems to be ancestral to most Mentha species that were retrieved from the database [Figure 3a].
In plants, the most commonly amplified and sequenced gene for taxonomic and phylogenetic studies is the plastid rbcL [79-81]. Hence, sequences of the rbcL gene from Madinah mint were obtained, and they were able to discriminate this species from other retrieved species, except that they were identical to sequences from M. canadensis and M. spicata [Table 2 and Figure 3b]. Plastid rbcL gene is well accepted in phylogenetic studies of plants, but delimiting a taxa to a single species based on rbcL gene alone is difficult, especially in closely related species [28,82-86] such as Mentha. Phylogenetic analysis using the Neighbor-Joining method and partial sequences from the rbcL gene revealed that Madinah mint is ancestral to M. suaveolens, M. canadensis, and M. spicata. Therefore, it is possible to say that the obtained partial sequences of matK and rbcL genes were fully capable of delimiting Madinah mint and can be applied to discriminate it from similar species due to these observed nucleotide differences.
The intergenic space trnA-trnH sequences from Madinah mint was not able to retrieve any similar Mentha species except for related taxa, probably due to a lack of similar sequences from Mentha species available in the database. Sequences of the plastid rpoC1 gene from Madinah mint showed 100% base identity with M. spicata, but 99.78% with M. aquatica [Figure 2]. Yet, this partial rpoC1 sequence can be applied as a DNA marker to differentiate between these two species, Madinah mint and M. aquatica based on detected SNPs. Alaklabi et al. [28] used SNPs found in a partial rbcL gene sequence to validate and differentiate D. mespiliformis tree. Sequences of the ycf5 gene showed that Madinah mint is similar to M. x piperita, M. spicata, and M. canadensis (98.92%). The ycf5 gene was applied in the study of inter/intra-specific variation of medicinal plant species [53].
In agreement with plastid loci above, the nuclear ITS1 and ITS2 were very successful and able to discriminate Madinah mint from all other Mentha species available on the database, despite the close relationships with some species. ITS1 and ITS2 can be applied as species-specific markers or DNA barcodes for the purpose of authentication and identification of this species of Mentha traded under the name Madinah mint in Saudi Arabia. The ITS region has been utilized as a DNA barcode to help identify over 21,000 plant species [51].
Phylogenetic analyses utilizing various genetic markers have provided insights into the evolutionary relationships of Madinah mint. The matK gene analysis positioned Madinah mint as a basal lineage to other Mentha species, excluding M. cervina [Figure 3a]. Conversely, rbcL sequence analysis placed Madinah mint as ancestral to M. canadensis, M. spicata, and M. suaveolens, while sharing a common origin with M. aquatica and M. longifolia [Figure 3b]. Combining both matK and rbcL data positioned Madinah mint as ancestral to all species except M. longifolia [Figure 3c]. When ITS1 or ITS2 sequences were used, Madinah mint appeared as ancestral to or a sister species of M. canadensis and M. spicata [Figure 4a and b]. However, the combined sequence analysis aligned with the results from matK and rbcL, positioning Madinah mint as a basal taxon to other species [Figures 3a-c and 4c]. This pattern is likely due to the shared origin of maternally inherited plastid genomes among these species [47].
5. CONCLUSION
Based on sequences alignment and phylogenetic analysis, this study demonstrated a very close relationship between Madinah mint and other related Mentha species based on at least partial coding sequences from two plastid genes (matK and rbcL) and two non-coding partial sequences from the nucleus (ITSs). The present findings showed that partial sequences of matK and rbcL genes were able to delimit the Madinah mint to its species level. The partial sequences obtained from the trnA-trnH intergenic space were not able to retrieve similar Mentha species probably due to lack of these sequences in the database. The sequences from ycf5 gene differentiated Madinah mint from M. x piperita, M. spicata and M. canadensis. However, sequences from the nuclear ITS1 and ITS2 were able to position Madinah mint close to M. spicata and M. canadensis, yet, there were some differences. Therefore, it is recommended to use SNPs detected during this study in sequences of matk and rbcL genes and/or ITSs to authenticate and identify Madinah mint that is mixed in trade with other related species. Phylogenetic analysis using the Neighbor-Joining method and sequences from the rbcL gene were able to evolutionary position Madinah mint to a distinct phylogenetic clade with M. suaveolens, M. spicata, and M. canadensis, which may suggest that these three species have shared the same origin of the maternally inherited plastid genome.
6. ACKNOWLEDGMENTS
The author would like to acknowledge the support from the Department of Biological Sciences, College of Science, King Faisal University, Saudi Arabia. He would also like to thank those anonymous reviewers for their useful comments and suggestions.
7. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
8. FUNDING
There is no funding to report.
9. CONFLICTS OF INTEREST
The author report no financial or any other conflicts of interest in this work.
10. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
11. DATA AVAILABILITY
Most of the data generated and analyzed are included in this research article. If required, more data can be obtained from the corresponding author, Assistant Professor Rashid IH Ibrahim, via email at [email protected].
12. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
13. Use of artificial intelligence (AI)-assisted technology
The authors declare that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Singh G. Plant Systematics: An Integrated Approach. New Hampshire: Science Publishers;2010.
2. Assaf M, Korkmaz A, Karaman S, Kulak M. Effect of plant growth regulators and salt stress on secondary metabolite composition in Lamiaceae species. South Afr J Bot. 2022;144:480-93.[CrossRef]
3. Naghibi F, Mosaddegh M, Motamed SM, Ghorbani A. Labiatae family in folk medicine in Iran:From ethnobotany to pharmacology. Iran J Pharm Res. 2005;2:63-79.
4. Barros L, Heleno SA, Carvalho AM, Ferreira IC. Lamiaceae often used in Portuguese folk medicine as a source of powerful antioxidants:Vitamins and phenolics. LWT. 2010;43:544-50.[CrossRef]
5. Djilani A, Dicko A. The Therapeutic benefits of essential oils. In:Nutrition, Well-Being and Health. London:InTechopen;2012.[CrossRef]
6. Nazar N, Howard C, Slater A, Sgamma T. Challenges in medicinal and aromatic plants DNA barcoding-lessons from the Lamiaceae. Plants (Basel). 2022;11:137.[CrossRef]
7. Ramasubramania Raja R. Medicinally potential plants of labiatae (Lamiaceae) family:An overview. J Med Plant. 2012;3:203-13.[CrossRef]
8. Dorman HJ, Peltoketo A, Hiltunen R, Tikkanen MJ. Characterisation of the antioxidant properties of de-odourised aqueous extracts from selected Lamiaceae herbs. Food Chem. 2003;83:255-62.[CrossRef]
9. Matkowski A, Piotrowska M. Antioxidant and free radical scavenging activities of some medicinal plants from the Lamiaceae. Fitoterapia. 2006;77:346-53.[CrossRef]
10. De Mattia F, Bruni I, Galimberti A, Cattaneo F, Casiraghi M, Labra M. A comparative study of different DNA barcoding markers for the identification of some members of Lamiacaea. Food Res Int. 2011;44:693-702.[CrossRef]
11. Theodoridis S, Stefanaki A, Tezcan M, Aki C, Kokkini S, Vlachonasios KE. DNA barcoding in native plants of the Labiatae (Lamiaceae) family from Chios Island (Greece) and the adjacent Çesme-Karaburun Peninsula (Turkey). Mol Ecol Resour. 2012;12:620-33.[CrossRef]
12. Celep F, Dirmenci T. Systematic and biogeographic overview of Lamiaceae in Turkey. Nat Volatiles Essential Oils. 2017;4(4):14-27.
13. Zhao F, Chen YP, Salmaki Y, Drew BT, Wilson TC, Scheen AC, et al. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 2021;19:2.[CrossRef]
14. Hussin N, Mohd Noor NN, Mohamed F. Taxonomic significance of trichome ultrastructure in five genera of Lamiaceae. J Sci Math Lett. 2024;12:8-17.[CrossRef]
15. Brown N, John JA, Shahidi F. Polyphenol composition and antioxidant potential of mint leaves. Food Prod Process Nutr. 2019;1:1.[CrossRef]
16. Al-Ali K, Abdelrazik M, Alghaithy A, Diab A, El-Beshbishy H, Baghdadi H. Antimutagenic and anticancer activity of Al Madinah Alhasawy mint (Mentha longifolia) leaves extract. Pakistan Journal of Biological Sciences 17(12):1231-6. doi:10.3923/pjbs.2014.1231.1236[CrossRef]
17. Tafrihi M, Imran M, Tufail T, Gondal TA, Caruso G, Sharma S, et al. The wonderful activities of the genus Mentha:Not only antioxidant properties. Molecules. 2021;26:1118.[CrossRef]
18. Hebert PD, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proc R Soc B Biol Sci. 2003;270:313-21.[CrossRef]
19. Abdel-Hamid AM, Elenazy HH, Abdel-Hameed UK. DNA barcoding of some taxa of genus Acacia and their phylogenetic relationship. All Life. 2021;14:588-98.[CrossRef]
20. Jamdade R, Mosa KA, El-Keblawy A, Al Shaer K, Al Harthi E, Al Sallani M, et al. DNA Barcodes for accurate identification of selected medicinal plants (Caryophyllales):Toward barcoding flowering plants of the United Arab Emirates. Diversity (Basel). 2022;14:262.[CrossRef]
21. Abdelaziz SA, Khaled KA, Younis RA, Al-Kordy MA, El-Domyati FM, Moghazee MM. Comparison of four DNA barcoding loci to distinguish between some Apiaceae family species. Beni Suef Univ J Basic Appl Sci. 2024;13:12.[CrossRef]
22. Lonare N, Patil G, Waghmare S, Bhor R, Hardikar H, Tembe S. DNA barcoding of invasive terrestrial plant species in India. Mol Biotechnol. 2024;67(3):1027-1034. doi:10.1007/s12033-024-01102-z[CrossRef]
23. Lefébure T, Douady CJ, Gouy M, Gibert J. Relationship between morphological taxonomy and molecular divergence within Crustacea:Proposal of a molecular threshold to help species delimitation. Mol Phylogenet Evol. 2006;40:435-47.[CrossRef]
24. Wong EH, Hanner RH. DNA barcoding detects market substitution in North American seafood. Food Res Int. 2008;41:828-37.[CrossRef]
25. Frézal L, Leblois R. Four years of DNA barcoding:Current advances and prospects. Infect Genet Evol. 2008;8:727-36.[CrossRef]
26. Bruni I, De Mattia F, Galimberti A, Galasso G, Banfi E, Casiraghi M, et al. Identification of poisonous plants by DNA barcoding approach. Int J Legal Med. 2010;124:595-603.[CrossRef]
27. Bafeel SO, Alaklabi A, Arif IA, Khan HA, Alfarhan AH, Ahamed A, et al. Ribulose-1, 5-biphosphate carboxylase (rbcL) gene sequence and random amplification of polymorphic DNA (RAPD) profile of regionally endangered tree species Coptosperma graveolens subsp. (S. Moore) Degreef. Plant Omics. 2012;5:285-90.
28. Alaklabi A, Arif IA, Bafeel SO, Alfarhan AH, Ahamed A, Thomas J, et al. Nucleotide based validation of the endangered plant Diospyros mespiliformis (Ebenaceae) by evaluating short sequence region of plastid rbcL gene. Plant Omics. 2014;7:102-7.
29. Safhi FA, Alshamrani SM, Bogmaza AF, El-Moneim DA. DNA barcoding of wild plants with potential medicinal properties from faifa mountains in Saudi Arabia. Genes (Basel). 2023;14:469.[CrossRef]
30. Han JP, Shi LC, Chen XC, Lin YL. Comparison of four DNA barcodes in identifying certain medicinal plants of Lamiaceae. J Syst Evol. 2012;50:227-34.[CrossRef]
31. Bayley A. A summary of current DNA methods for herb and SPICE identification. J AOAC Int. 2019;102:386-9.[CrossRef]
32. Sasikumar B, Syamkumar S, Remya R, Zachariah TJ. PCR based detection of adulteration in the market samples of turmeric powder. Food Biotechnol. 2004;18:299-306.[CrossRef]
33. Dhanya K, Sasikumar B. Molecular marker based adulteration detection in traded food and agricultural commodities of plant origin with special reference to spices. Curr Trends Biotechnol Pharm. 2010;4:454-89.
34. Focke F, Haase I, Fischer M. DNA-based identification of spices:DNA isolation, whole genome amplification, and polymerase Chain reaction. J Agric Food Chem. 2011;59:513-20.[CrossRef]
35. Parvathy VA, Swetha VP, Sheeja TE, Leela NK, Chempakam B, Sasikumar B. DNA barcoding to detect chilli adulteration in traded black pepper powder. Food Biotechnol. 2014;28:25-40.[CrossRef]
36. Galimberti A, Labra M, Sandionigi A, Bruno A, Mezzasalma V, De Mattia F. DNA barcoding for minor crops and food traceability. Adv Agric. 2014;2014:831875.[CrossRef]
37. Thakur VV, Tripathi N, Tiwari S. DNA barcoding of some medicinally important plant species of Lamiaceae family in India. Mol Biol Rep. 2021;48:3097-106.[CrossRef]
38. Raclariu-Manolica AC, Anmarkrud JA, Kierczak M, Rafati N, Thorbek BL, Schrøder-Nielsen A, et al. DNA metabarcoding for quality control of Basil, Oregano, and Paprika. Front Plant Sci. 2021;12:3097-106.[CrossRef]
39. Waugh J. DNA barcoding in animal species:Progress, potential and pitfalls. BioEssays. 2007;29:188-97.[CrossRef]
40. Ratnasingham S, Hebert PD. A DNA-based registry for all animal species:The barcode index number (BIN) system. PLoS One. 2013;8:66213.[CrossRef]
41. Gorini T, Mezzasalma V, Deligia M, De Mattia F, Campone L, Labra M, et al. Check your shopping cart:DNA barcoding and mini-barcoding for food authentication. Foods. 2023;12:2392.[CrossRef]
42. Newmaster SG, Fazekas AJ, Ragupathy S. DNA barcoding in land plants:Evaluation of rbcL in a multigene tiered approach. Can J Bot. 2006;84:335-41.[CrossRef]
43. Hollingsworth ML, Andra Clark A, Forrest LL, Richardson J, Pennington RT, Long DG, et al. Selecting barcoding loci for plants:Evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour. 2009;9:439-57.[CrossRef]
44. CBOL Plant Working Group. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009;106:12794-7.[CrossRef]
45. Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, et al. Land plants and DNA barcodes:Short-term and long-term goals. Philos Trans R Soc B Biol Sci. 2005;360:1889-95.[CrossRef]
46. Chase MW, Cowan RS, Hollingsworth PM, Van Den Berg C, Madriñán S, Petersen G, et al.Aproposal for a standardised protocol to barcode all land plants. Taxon. 2007;56:295-9.[CrossRef]
47. Chase MW, Fay MF. Barcoding of plants and fungi. Science (1979). 2009;325:682-3.[CrossRef]
48. Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005;102:8369-74.[CrossRef]
49. Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants:The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007;2:508.[CrossRef]
50. Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, et al. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One. 2008;3:2802.[CrossRef]
51. Letsiou S, Madesis P, Vasdekis E, Montemurro C, Grigoriou ME, Skavdis G, et al. DNA barcoding as a plant identification method. Appl Sci (Switzerland). 2024;14:1415.[CrossRef]
52. Filipowicz N, Nee M, Renner S. Description and molecular diagnosis of a new species of Brunfelsia (Solanaceae) from the Bolivian and Argentinean Andes. PhytoKeys. 2012;10:83.[CrossRef]
53. Chen S, Yao H, Han J, Liu C, Song J, Shi L, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613.[CrossRef]
54. Saarela JM, Sokoloff PC, Gillespie LJ, Consaul LL, Bull RD. DNA barcoding the canadian arctic flora:Core plastid barcodes (rbcL +matK) for 490 vascular plant Species. PLoS One. 2013;8:77982.[CrossRef]
55. Tanabe AS, Toju H. Two new computational methods for universal DNA barcoding:A benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS One. 2013;8:76910.[CrossRef]
56. Fu N, Xu Y, Jin L, Xiao TW, Song F, Yan HF, et al. Testing plastomes and nuclear ribosomal DNA sequences as the next-generation DNA barcodes for species identification and phylogenetic analysis in Acer. BMC Plant Biol. 2024;24:445.[CrossRef]
57. Altschup SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-10.[CrossRef]
58. Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947-8.[CrossRef]
59. Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247-50.[CrossRef]
60. Tamura K, Stecher G, Kumar S. MEGA11:Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022-7.[CrossRef]
61. Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125:1-15.[CrossRef]
62. Ross HA, Murugan S, Li WL. Testing the reliability of genetic methods of species identification via simulation. Syst Biol. 2008;57:216-30.[CrossRef]
63. Kuzmina ML, Johnson KL, Barron HR, Hebert PD. Identification of the vascular plants of Churchill, Manitoba, using a DNA barcode library. BMC Ecol. 2012;12:25.[CrossRef]
64. Syme AE, Udovicic F, Stajsic V, Murphy DJ. A test of sequence-matching algorithms for a DNA barcode database of invasive grasses. DNA Barcodes. 2013;1:19-26.[CrossRef]
65. Tsaballa A, Kelesidis G, Krigas N, Sarropoulou V, Bagatzounis P, Grigoriadou K. Taxonomic identification and molecular DNA barcoding of collected wild-growing orchids used traditionally for salep production. Plants (Basel). 2023;12:3038.[CrossRef]
66. Lanubile A, Stagnati L, Marocco A, Busconi M. DNA-based techniques to check quality and authenticity of food, feed and medicinal products of plant origin:A review. Trends Food Sci Technol. 2024;149:104568.[CrossRef]
67. Jamdade R, Upadhyay M, Al Shaer K, Al Harthi E, Al Sallani M, Al Jasmi M, et al. Evaluation of arabian vascular plant barcodes (rbcL and matK):Precision of unsupervised and supervised learning methods towards accurate identification. Plants. 2021;10:2741.[CrossRef]
68. Algarni AA. Evaluation of plastid and nuclear DNA markers in barcoding of Aloe saudiarabica, KSA. Cell Mol Biol (Noisy-le-Grand). 2023;69:126-32.[CrossRef]
69. De Boer H, Orwick M, Verstraete RB, Gravendeel B. Molecular Identification of Plants:From Sequence to Species. Prof. Georgi Zlatarski Str. 12 1111. Sofia, Bulgaria:Pensoft;2022.[CrossRef]
70. Xiong C, Sun W, Wu L, Xu R, Zhang Y, Zhu W, et al. Evaluation of four commonly used DNA barcoding loci for Ardisia species identification. Front Plant Sci. 2022;13:860778.[CrossRef]
71. Wei L, Pacheco-Reyes FC, Villarreal-Quintanilla JÁ, Robledo-Torres V, Encina-Domínguez JA, Lara-Ramírez EE, et al. Effectivness of DNA barcodes (rbcL, matK, ITS2) In identifying genera and species in Cactaceae. Pak J Bot. 2024;56:1911-28.[CrossRef]
72. Bafeel SO, Arif IA, Bakir M, Khan H. Comparative evaluation of PCR success with universal primers of maturase K (matK) and ribulose-1, 5-bisphosphate carboxylase oxygenase large subunit (rbcL) for barcoding of some ar. Plant Omics. 2011;4:195-8.
73. Hollingsworth PM. Refining the DNA barcode for land plants. Proc Natl Acad Sci U S A. 2011;108:19451-2.[CrossRef]
74. Yu J, Xue JH, Zhou SL. New universal matK primers for DNA barcoding angiosperms. J Syst Evol. 2011;49:176-81.[CrossRef]
75. Karbarz M, Szlachcikowska D, Zapal A, Lesko A. Unlocking the genetic identity of endangered Paphiopedilum orchids:A DNA barcoding approach. Genes (Basel). 2024;15:689.[CrossRef]
76. Nair RR, Udayan PS, Thilaga S, Kavitha M, Bharathanandhini RM, Nizzy AM, et al. Molecular distinction of two closely resembling Morinda species using rbcL and matK loci for quality management of Indian herbal medicines. Plant Genet Resour Character Util. 2013;11:90-3.[CrossRef]
77. Bharatha Nandhini RM, Rahul RN, Thilaga S, Rao NS, Ganesh D. Molecular distinction of C×R hybrid (Coffea congensis×Coffea canephora) from morphologically resembling male parent using rbcL and matK gene sequences. South Afr J Bot. 2013;88:334-40.[CrossRef]
78. Wong KH, Zheng T, Yue GG, Li MC, Wu HY, Tong MH, et al.Asystematic approach for authentication of medicinal Patrinia species using an integration of morphological, chemical and molecular methods. Sci Rep. 2024;14:6566.[CrossRef]
79. Schuettpelz E, Korall P, Pryer KM. Plastid atpA data provide improved support for deep relationships among ferns. Taxon. 2006;55:897-906.[CrossRef]
80. Zhu S, Liu Q, Qiu S, Dai J, Gao X. DNA barcoding:An efficient technology to authenticate plant species of traditional Chinese medicine and recent advances. Chin Med. 2022;17:112.[CrossRef]
81. Yong WTL, Mustafa AA, Derise MR, Rodrigues KF. DNA barcoding using chloroplast matK and rbcL regions for the identification of bamboo species in Sabah. Adv Bamboo Sci. 2024;7:100073.[CrossRef]
82. Gadek PA, Quinn CJ. An analysis of relationships within the Cupressaceae sensu stricto based on rbcL sequences. Ann Missouri Bot Garden. 1993;80:581-6.[CrossRef]
83. Les DH, Philbrick CT, Novelo A. The phylogenetic position of river-weeds (Podostemaceae) Insights from rbcL sequence data. Aquat Bot. 1997;57:5-27.[CrossRef]
84. Chen ZD, Manchester SR, Sun HY. Phylogeny and evolution of the Betulaceaeas inferred from DNA sequences. Am J Bot. 1999;86:1168-81.[CrossRef]
85. Weerasena J, Rajphriyadharshini R, Weerasena OV. DNA barcoding of medicinal plant:A systemic review. 2020;9:6-16.
86. Rattray RD, Stewart RD, Niemann HJ, Olaniyan OD, van der Bank M. Leafing through genetic barcodes:An assessment of 14 years of plant DNA barcoding in South Africa. South Afr J Bot. 2024;172:474-87.[CrossRef]
87. Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406-25.
88. Nei M, Kumar S. Molecular Evolution and Phylogenetics. Madison Avenue, New York, New York 10016, USA:Oxford University Press, Inc.198;2000.