1. INTRODUCTION
Stephania is the largest genus of the family Menispermaceae with about 60 species distributed in the tropical and subtropical regions of Asia and Africa. Some species are also found in Oceania. Recently, 37 species were recorded in China and 15 species in Thailand [1–3]. Because their tubers contain a number of important alkaloids, such as L-tetrahydropalmatine (rotundin), stepharin, roemerin, and cycleanin, Stephania spp. have long been used in traditional medicine to treat various diseases such as sedation, blood pressure stabilization, asthma, tuberculosis, dysentery, hyperglycemia, malaria, and cancer [4–6].
In Vietnam, this genus comprises 20 species with the similar dioecious flower [4], including several medicinal species such as Stephania cepharantha Hayata, Stephania rotunda Lour., and Stephania japonica Miers which are being overexploited and listed in the Red Data Book of Vietnam with a level of “going to be endangered” (V). However, the conservation of genetic resources of the species in the genus Stephania is still difficult due to the misidentification of species with similar morphological and anatomical characteristics.
Plant species can be identified by many different analytical methods. The current methods such as analysis and comparison of morphological, anatomical, physiological, or biochemical characteristics have been reported successfully in a number of crops such as Fallopia multiflora (Thunb.) Haraldson [7], Albizia myriophylla Benth. [8], and Pelargonium hortorum L. H. Bailey [9]. However, it is difficult to efficiently identify plants, especially closely related species which belong to the same subgenus or their parts are not intact.
Recently, DNA barcode data have been widely and regularly used to provide additional evidence at the molecular level for plant taxonomic studies. The trend of combining morphological characteristics and chemical and genetic markers into a dataset for species identification becomes very important for systematic studies, in which DNA barcoding has become one of the most efficient tools for species identification of medicinal plants. Several barcoding loci including matK, rpoC1, trnH-psbA, ITS, and rbcL have been studied and applied effectively in the identification of medicinal plants [10–12]. The matK gene found in chloroplasts has been successfully applied to plant identification [11]. The ITS gene region located in the cell nucleus, including the ITS1-5.8S-ITS2 sequence, has achieved high identification rates at the species level. The studies determining the phylogenetic relationships between plant species based on ITS genome sequencing in Dalbergia tonkinensis, Dalbergia cochinchinensis, and Dalbergia oliveri [13] or genera Erica L. [14], Scrophularia [15], and Potamogeton [16] and many other plant species demonstrate the role of the ITS gene region in plant identification [17,18]. In this study, the Stephania brachyandra collected in Lào Cai (Vietnam) was identified using the comparative morphological method and supported the DNA barcode method with matK and ITS.
2. MATERIALS AND METHODS
2.1. Materials
Stephania spp. samples were collected in Lào Cai province, Vietnam, and these samples were classified at the Laboratory of Botany and Genetics and grown in the Experimental Garden at the School of Biology, Thai Nguyen University of Education, Vietnam.
2.2. Morphological Analysis
The morphological features of Stephania spp. were studied following the protocol of Nguyen et al. [19], Flora of Vietnam by Pham [20], and Paris Linnaeus by Liang and Soukup [21]. Key indicators used for analysis include the height of the main stem and stem color; the number of leaves, leaf shape, and leaf size; and the shape, color, and number of calyxes, corollas, stamens, and pistils.
2.3. DNA Extraction, Polymerase Chain Reaction (PCR) Amplification, and Sequencing
Total genomic DNA was extracted from young fresh leaves material following the protocol of Shaghai-Maroof et al. [22]. The sequences of the matK gene and ITS region in Stephania spp. plants were amplified by PCR using the primer pairs presented in Table 1.
 | Table 1: Characteristics of matK and ITS primer pairs for the PCR. [Click here to view] |
The PCR amplification was carried out using a final volume of 25 μl with 1.5 μl forward primers (10 pmol μl−1), 1.5 μl reverse primers (10 pmol μl−1), 12.5 μl 2× Master Mix, 1.0 μl template genomic DNA (500 ng ml−1), and 8.5 μl deionized water. The PCR amplification profiles consisted of 4 minutes at 94°C for initial denaturation, 30 cycles of 1 minute at 94°C, annealing for 1 minute at 54°C, 1 minute 30 seconds at 72°C for extension, and a final extension step for 10 minutes at 72°C. The PCR products were detected by 1.0% agarose gel electrophoresis.
The matK and ITS sequences were identified by the machine ABI PRISM® 3100 Avant Genetic Analyzer with Kit BigDye® Terminator v3.1 Cycle Sequencing and a specific primer pair. The data were analyzed by the basic local alignment search tool (BLAST) tool.
2.4. Phylogenetic Analysis
The evolutionary history was inferred by using the maximum likelihood (ML) method and the Tamura and Nei model [23]. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed [24]. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using the maximum composite likelihood approach. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated (complete deletion option). Evolutionary analyses were conducted in MEGA X [25].
3. RESULTS
3.1. Plant Sample Collection and Morphological Identification of S. brachyandra
The Stephania sample collected in Lào Cai (Vietnam) is perennial with herbaceous climbing vines up to 2–3 m long and a woody stem base; stem, leaves, and flowers are usually hairless (Fig. 1). The analysis of morphological characteristics of the collected Stephania_Laocai has indicated that this species is S. brachyandra Diels. It is a simple and alternate leaf, and petioles about 5–10 cm long are attached to the leaf blade at about one-third to one-sixth of the leaf length. The leaf blade is thin, glabrous, and egg- shaped to triangular or rounded, 515 cm, with the entire margins being smooth, base flat, or slightly convex. The leaf veins are propeller- shaped, consisting of 8–11 veins originating from the top of the petiole. The leaf tip is pointed or nearly rounded; the leaf base is rounded or heart- shaped. The front side of the leaf blade is dark green, while a pale green or slightly silver color is seen at the backside.
 | Figure 1: The morphology of S. brachyandra Diels collected in Lào Cai, Vietnam. (A) S. brachyandra plants; (B and C) upperside and underside of leaves, respectively; (D): stem and tuberous tuber root; (E): flowers; (G): fruits; and (H): cross-section of fruit. Scale bar: 1.0 mm. [Click here to view] |
Shoots and young stems are usually smooth, dark green, light green, or glossy green. The outer layer of the bark has cracks along the stem or rough warts. The stems are ash gray, dark brown, and light brown. Tuberous roots are very diverse, usually spherical, lean, and brown. The size and weight of the tuber are very different, ranging from about 1–3 kg up to 80 kg. The inner part of the tuber is light yellow or lemon yellow, ivory white, or reddish-brown.
The flowers are unisexual, with the inflorescences compound umbelliform cymes, double canopy, single canopy, or head- shaped [26]; the inflorescences have peduncles, solitary or clustered on the primary inflorescence branches; the terminal branches sometimes irregular or the cymes gather into a disk that is head- shaped [27]. Male inflorescences are slightly slender: peduncle 2–4 cm, six sepals arranged in two rings, three yellow-orange petals,m and disk-shaped anthers. Female inflorescences have shorter stalks than the male inflorescences because of 8–9 small cymes, tightly arranged in a headed shape. Inflorescence peduncle is 2–3 cm, with the apex slightly swollen. Flowers are densely arranged. Therefore, it is hard to see the flower stalk. Female flowers are usually small: sepal one, pale green; two petals arranged on the same side of the flower, yellow-orange, with the shape of an inverted ovate. The ovary is ovate and curved, with a short peduncle; the stigma has –four to five small spiny lobes. Flowers are cross-pollinated mainly by several insect species [27,28].
The fruit has only one seed, 0.7–0.8 cm, ovate to nearly round, flattened on both sides. The outer skin is usually orange-red, smooth, and shiny when ripe. The ovary has two ovules, but only one develops into the seed, whereas the other one degenerates. Seeds are horseshoe-shaped, inverted ovate, amputated heads, membranous connected to the semicircular ring; in the middle of the seeds, there is an inverted ovoid hole. Along the dorsal and ventral edge of the seed, there are four rows of spines with bulging heads that swell up into the shape of a nail cap [26].
3.2. Analysis of matK and ITS Sequences of S. brachyandra
3.2.1. Total gGenomic DNA eExtraction and PCR aAmplification of the matK gGene and ITS rRegion
The purification of total genomic DNA extracted from leaves tissues of S. brachyandra was assessed via agarose gel electrophoresis and measured using a spectrophotometer. The result showed that the specific band was clean and had no contamination of RNA and protein (data not shown). The matK gene and ITS region of the genomic DNA were amplified by PCR using primer pairs matK-F/matK-R and ITS-F/ITS-R, respectively. The PCR products detected by 1.0% agarose gel electrophoresis revealed a DNA fragment of the matK gene and ITS region with the expected sizes of approximately 900 and 500 bp, respectively (Fig. 2).
The PCR products of the matK and ITS sequence were purified and sequenced on an ABI PRISM® 3100 automated sequencer, and the results showed that matK is 879 bp in size and ITS is 423 bp in size. The BLAST analysis showed that the matK and ITS sequences of Stephania_Laocai were close to S. brachyandra, and the matK sequence provided for 74% of query coverage and showed relatedness to S. brachyandra with 879 nucleotides of a total BLAST score and with a 99.37% sequence identity and the ITS provided for 100% of query coverage and showed relatedness to S. brachyandra with 423 nucleotides of a total BLAST score and with a 98.97% sequence identity.
The results of comparing 28 matK sequences by BLAST showed that the isolated matK sequence was close to species of the Stephania genus and provided for 53%–74% of query coverage and relatedness to the Stephania species with 342–571 of a total BLAST score and with 86.35%–99.37% sequence identity (Table 2). The aligned sequence of the matK gene showed 879 bp length, and the matK sequences were highly conserved and had low variable sites for 747 nucleotides (84.98%) and 132 nucleotides (15.29%), respectively.The results of comparing 20 ITS sequences by BLAST showed that the isolated ITS sequence was close to 13 species of the Stephania genus and seven species of the other genus and provided for 100% of query coverage and relatedness to the Stephania species with 1,378–1,567 of a total BLAST score and with 95.10%–98.97% sequence identity (Table 3). For the short ITS region, the aligned sequence showed 423 bp length and was less conserved and had variable sites for 78 (18.44%) and 345 (81.56%), respectively.
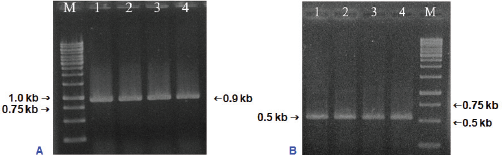 | Figure 2: PCR amplification of the matK gene (A) and ITS region (B). M: Marker 1 kb; 1–4: PCR products of matK/ITS. [Click here to view] |
 | Table 2: Twenty-eight species in the top 100 BLAST hits of matK. [Click here to view] |
3.3. Phylogenetic Analysis
Recently, the ML method has been applied for DNA sequence analysis [29]. The results of the molecular phylogenetic analysis of the matK gene by the ML method (Fig. 3) for the Stephania_Laocai sample showed the ML tree from the matK locus. DNA barcode alignment was carried out with the highest log likelihood (−2,148.48). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 20 nucleotide matK sequences including a Stephania_Laocai sample and 19 matK sequences in GenBank. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 862 positions in the final dataset. In Figure 3, 13 species of the Stephania genus are grouped in 1 clade, which is divided into 2 subclades with 99%–100% support, in which the Stephania_Laocai sample and S. brachyandra (EF143871.1) [30] are in 1 subclade (bootstrap values = 76%). In contrast to the matK sequence, the ITS region dataset yielded less phylogenetic resolution than the bootstrap value which was 59% at the clade of the genus Stephania (Fig. 4). Additionally, the genus Stephania was separated into two main branches (bootstrap values = 99%). The second major branch further divides into two secondary branches and the second secondary branch divides into many clades and subclades. Thus, in the case of barcoding among species of the Stephania genus and to identify the S. brachyandra species, the matK sequence is suggested for better phylogenetic resolution.
Among the species of the genus Stephania, only a few species with both the matK gene and the ITS region sequences were found in GenBank. The phylogenetic tree was established based on the matK and ITS sequence combinations (matK/ITS) shown and the Stephania_Laocai and S. brachyandra samples distributed in a clade with a bootstrap value of 97% (Fig. 5). Thus, the combination of matK and ITS can be used to identify S. brachyandra.
4. DISCUSSION
Along with this approach, Chinh et al. [31] used the chloroplast rbcL gene to clarify the relationship between three species of the genus Stephania (Menispermaceae) from Vietnam, S. japonica, Stephania polygona, and S. rotunda, and one subspecies, S. japonica var. discolor. According to these authors, the rbcL gene of the chloroplast genome is widely used as additional data for the study of species origin, molecular evolution, and phylogeny. Molecular analysis was carried out on the 523 bp segment of the rbcL genes. The dataset consists of 22 sequences used to reconstruct the evolutionary tree using two methods: Bayesian inference and mlML. The results indicated that S. rotunda was able to distinguish between S. japonica and S. polygona, while S. japonica, S. japonica var. discolor, and S. polygona could not distinguish each other. However, the rbcL gene also has its limitations. Previous studies showed that 58.5% of sister species were not identifiable by the rbcL gene sequence because of 100% similarity [12]. Hence, they suggested that other loci such as nuclear ITS and chloroplast trnH-psbA space should be examined further or a combination of multiple gene loci for the genus Stephania should be studied. Wang et al. [29] confirmed that the genus Stephania is polyphyly, which has been grouped but does not share an immediate common ancestor based on phylogeny and morphological evolution of the tribe Menispermeae (Menispermaceae) inferred from chloroplast and nuclear sequences. The inconsistency between the molecular system and the traditional classification system has been pointed out in the genera of Menispermaceae [5]. Therefore, a combination of morphological and molecular characteristics is needed to rearrange the classification system of the Stephania genus. DNA barcodes (or molecular markers) are an effective tool to support morphology in species identification and rearrangement of the classification system.
 | Table 3: Twenty species in the top 100 BLAST hits of ITS. [Click here to view] |
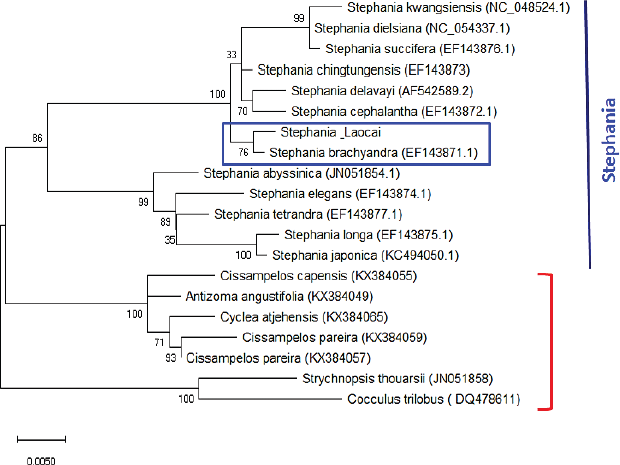 | Figure 3: Molecular phylogenetic analysis of the matK gene. The 20 matK sequences were obtained including a Stephania_Laocai sample and 19 matK sequences in GenBank, in which there are 13 matK sequences belonging to the Stephania genus. Bootstrap values are above the nodes of the branches. The capital letters and numbers in parentheses are accession numbers of Stephania species published in the GenBank. [Click here to view] |
 | Figure 4: Molecular phylogenetic analysis of the ITS region. The 28 ITS sequences were obtained including a Stephania_Laocai sample and 27 ITS sequences in the GenBank. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Codon positions included were 1st + 2nd + 3rd + noncoding. All positions containing gaps and missing data were eliminated (complete deletion option). There were a total of 250 positions in the final dataset. Bootstrap values are above the nodes of the branches. The capital letters and numbers in parentheses are accession numbers of the Stephania species published in the GenBank. [Click here to view] |
 | Figure 5: Phylogenetic tree of the Stephania species constructed on the matK combined with ITS (matK/ITS). Bootstrap values are above the nodes of the branches. [Click here to view] |
5. CONCLUSION
In this study, the morphological features of a Stephania_Laocai sample, as well as two DNA barcodes matK and ITS, were analyzed to identify this species. Our results demonstrate that the Stephania spp. sample collected in Lào Cai province, Vietnam, is S. brachyandra Diels and is proposed to use the matK gene or combine matK with ITS to identify S. brachyandra species.
6. ACKNOWLEDGMENT
This study was funded by the Project of Ministry of Education and Training under grant number B2019-TNA-09.
7. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
8. ETHICAL APPROVAL
This article does not contain any studies involving animals or human participants performed by any of the authors.
9. CONFLICT OF INTEREST
The authors declare that they have no conflicts of interest.
REFERENCES
1. Hu Q, Luo X, Chen T, Gilbert MG. Menispermaceae. Flora of China, 2008;7: 1,166
2. Vu TC, Bui HQ, Choudhary RK, Xia NH, Lee J. Stephania subpeltata H.S.Lo (Menispermaceae): a new recorded species from Vietnam. Korean J Pl Taxon 2016;46(3):288–94. CrossRef
3. Trinh TP, Vu TC, Tran VT, Nong VD. The Species of the genus Stephania Lour. (Menispermaceae) in flora of Vietnam. Vietnam J Forest Sci 2019;4:22?35.
4. Nguyen TH, Nguyen QH, Pham HTT, Hoang VT. Taxonomy of some species in the genus Stephania Lour. in Vietnam using rDNA ITS sequences. J Pharmacy 2014;54(9):55?9.
5. Xie DA, He JA, Huang JA, Xie HA, Wang YA, Kang YAC, et al. Molecular phylogeny of Chinese Stephania (Menispermaceae) and reassessment of the subgeneric and sectional classifications. Aust Syst Bot 2015;28:246–55. CrossRef
6. Vu TC, Nong VD, Tran VT, Xia NH. Stephania polygona (Menispermaceae): a new species from Southern Vietnam. Phytotaxa 2019;400(3):211?4. CrossRef
7. Pham TH, Nguyen QN, Phan VT, Hoang VT, Nguyen XN, Pham TN, et al. Studies on morphological and anatomical characteristics of Fallopia multiflora (Thunb.) Haraldson in Vietnam. Proceedings of the 6th National Scientific Conference on Ecology and Biological Resources, Publishing House of Natural Science and Technology, Hanoi, Vietnam, pp 166–72, 2015
8. Nong TTA, Nguyen QT, Nguyen TMT, Dao TH. Study on the phytomorphology of the Albizia myriophylla Benth. plants collected in Thai Nguyen, Vietnam. Vietnam J Sci Technol 2017;17(6):10–2.
9. Ta LMH, Pham VM, Ha VO, Chu TTH, Dao TTH, Do TH. Research on botanical characteristics of Geranium (Pelargonium hortorum L. H. Bailey). J Pharmacol 2016;56(6):73–6.
10. Fazekas AJ, Burgess KS, Kesanakurti PR. Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS One 2008;3:2802. CrossRef
11. Lahaye R, Van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci U S A 2008;105(8):2923–8. CrossRef
12. Kang Y, Deng Z, Zang R, Long W. DNA barcoding analysis and phylogenetic relationships of tree species in tropical cloud forests. Sci Rep 2017;7:12564. CrossRef
13. Duong VT, Nguyen QB, Dinh TP. The nuclear ITS nucleotide sequences and phylogenetic relationship of three valuable wood species in Vietnam: Dalbergia cochinchinensis, D. oliveri and D. tonkinensis. Proceedings of the 4th National Scientific Conference on Ecology and Biological Resources, Agriculture Publishing House, Hanoi, Vietnam, pp 1296–1300, 2011
14. Pirie M, Michael D, Oliver EGH, Bellstedt DU. A densely sampled ITS phylogeny of the cape flagship genus Erica L. suggests numerous shifts in floral macro-morphology. Mol Phylogenet Evol 2011;61(2):593–601. CrossRef
15. Scheunert A, Heubl G. Diversification of Scrophularia (Scrophulariaceae) in the Western Mediterranean and Macaronesia–phylogenetic relationships, reticulate evolution and biogeographic patterns. Mol Phylogenet Evol 2014;70:296–313. CrossRef
16. Warwick SI, Mummenhoff K, Sauder CA, Koch MA, Al-Shehbaz IA. Closing the gaps: phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Syst Evol 2010;285:209–32. CrossRef
17. Hodkinson TR, Chase MW, Lledó DM, Dolores M, Salamin N, Renvoize SA. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. J Plant Res 2002;115(5):381–92. CrossRef
18. Yang T, Zhang T, Guo Y, Liu X. Identification of hybrids in Potamogeton: incongruence between plastid and ITS regions solved by a novel barcoding marker PHYB. PLoS One 2016;11(11):e0166177. CrossRef
19. Nguyen QN, Pham TH, Phan VT, Hoang VT. Taxonomy of the genus Paris L. (Melanthiaceae) in Vietnam. J Biol 2016;38(3):333–9. CrossRef
20. Pham HH. Flora of Vietnam. Youth Publishing House (in Vietnamese), Ho Chi Minh City, Vietnam, 1999.
21. Liang S, Soukup VG. Paris Linnaeus. Flora of China, St. Louis, MO, pp 88–95, 2000.
22. Shaghai-Maroof MA, Soliman KM, Jorgensen RA. Ribosomal DNA sepacer-length polymorphism in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci U S A 1984;81:8014–9. CrossRef
23. Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 1993;10:512–26.
24. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985;39:783–91. CrossRef
25. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35:1547–9. CrossRef
26. Bui KL, Pham TK. Contribution to the morphological study of Stephania species in Nghia Binh. J Pharmacol 1998;5:4–5.
27. Do TL. Medicinal plants and drugs from Vietnam. Medical Publising House, Hanoi, Vietnam, 2006.
28. Vu DT. Collection of Stephania spp. tubers in some provinces of Vietnam and quantification of rotundin using TLC-scanning. J Med Mater 2014;4:375–80.
29. Wang W, Wang HC, Chen ZD. Phylogeny and morphological evolution of tribe Menispermeae (Menispermaceae) inferred from chloroplast and nuclear sequences. Perspectives in plant ecology. Evol Syst 2007;8:141–54. CrossRef
30. Wang W, Wang HC., Chen ZD. Phylogeny and morphological evolution of tribe Menispermeae (Menispermaceae) inferred from chloroplast and nuclear sequences. Perspect Plant Ecol Evol Syst 2007; 8:141–54. CrossRef
31. Chinh VT, Lieu TT, Tang DV. Using the chloroplast rbcL gene to clarify the relationship between species of the genus Stephania (Menispermaceae) from Vietnam. Acad J Biol 2020;42(2):109–15. CrossRef