1. INTRODUCTION
Cancers pose a serious burden to global public health, with the number of cases and associated deaths rising annually [1]. Radiotherapy (RT) has been shown to be an effective palliative strategy and treatment as it increases longevity with disease-free survival in the affected. During RT, the ionizing radiation (IR) such as X-ray for multiple doses is delivered in fractions either via the conventional or fractionated regime. Radio-resistance refers to the ability of cancer cells to resist the cell-killing effects of radiation, thereby leading to reduced/compromised efficacy. Thus, the effectiveness of RT remains a challenge due to the development of resistance followed by recurrence and relapse even after the completion of therapy among a considerable number of breast cancer patients [2]. While therapeutic effects are mediated through direct DNA damage and cell killing, the resistance has been postulated to be mediated by non-targeted effects. One such phenomenon of non-targeted effect of IR is the adaptive response, specifically known as the radiation-induced adaptive response (RIAR) implicated in the development of radio-resistance. The low dose followed by high doses of IR serves to confer a radio-protective effect in normal cells whereas it reduces the therapeutic efficacy in cancer cells [3].
In vitro laboratory experiments with cell lines have demonstrated the existence of RIAR phenomenon followed by exposure to radiochemicals and radiation [4,5]. Various types of end-points (cytogenetic, genetic, proteomic, metabolic, or epigenetic) were employed to demonstrate the radio-resistance in those systems [6]. Scrutiny of literature has shown that many genes and gene products are involved in mediating the response; identifying the ideal candidate and inhibiting the molecule/pathway are proposed as new targets for cancer RT [7,8]. There are a lot of challenges in the existing therapeutic targets because of their specificity, sensitivity, and lack of standardization. Hence, an advancement in the categorization of genetic and epigenetic biomarkers via the in silico approaches may be helpful in identifying the pathways and molecules in mediating the response and confers resistance. The molecular mechanisms of RIAR have not been completely understood till date as this phenomenon remains challenging. Thus, understanding the gene-gene interactions is essential for unraveling the complex biological mechanisms in RIAR.
Evaluating the microRNAs (miRNAs) can serve as potential therapeutic targets to predict the RT response as they are profound theragnostic biomarkers [9]. Many miRNAs regulate the expression of radiation and cancer-related pathways; hence, targeting specific miRNAs of the target genes involved particularly in cellular pathways/mechanisms that mediate RIAR, which alter gene expression patterns during RT thereby paving the way for new possibilities in altering RIAR. Performing in silico studies can identify suitable targets that can pave the way for opting genes and miRNAs for future in vitro studies. Thereby, more appropriate studies can be performed to target miRNAs and alter gene expression during cancer RT. Therefore, we hypothesize that in silico analyses could reveal detecting and targeting the possible miRNAs associated with genes that might be potentially involved/mediated in the pathways of RIAR-associated mechanisms. The reason for the choice of breast tissue selected as the primary basis in this study for exploring RIAR is due to the possibilities of higher radio-resistance occurring in these types of cancers and an effective radiation therapy for cancer patients [10]. The objective of this in silico study is to find out the miRNAs that target the genes playing a crucial role in pathways associated with RIAR, using literature databases, integrating the gene set enrichment analysis (GSEA), and gene-gene interactions to predict the most possibly occurring miRNAs in RIAR and breast cancer. miRNAs function as post-transcriptional regulators by binding to messenger RNA and either inhibit the translation or stimulate degradation. Enriching miRNAs between the RIAR and breast cancer might help in finding the most appropriate miRNAs that will target genes altering the RIAR phenomenon during breast cancer RT. Though the studies have shown the role of miRNAs in mediating cell-cell communication and intercellular signal transduction for the occurrence of RIAR in fibroblasts as a protective mechanism, however, the knowledge of miRNAs in RIAR during clinical RT remains limited [7]. Therefore, this study remains novel in miRNA target prediction for the occurrence of RIAR phenomenon during breast cancer RT. The final miRNAs listed in this study can be further explored for in vitro experimental models as it can be candidate targets for future validation, it may be beneficial for altering the RT regimes (conventional or hyper-fractionated RT) in the clinic for enhanced therapeutic efficacy in breast cancer patients. Here, we provide detailed information on the rationale of gene interactions and highly enriched miRNA in breast cancer-associated genes that play a vital role in RIAR.
4. DISCUSSION
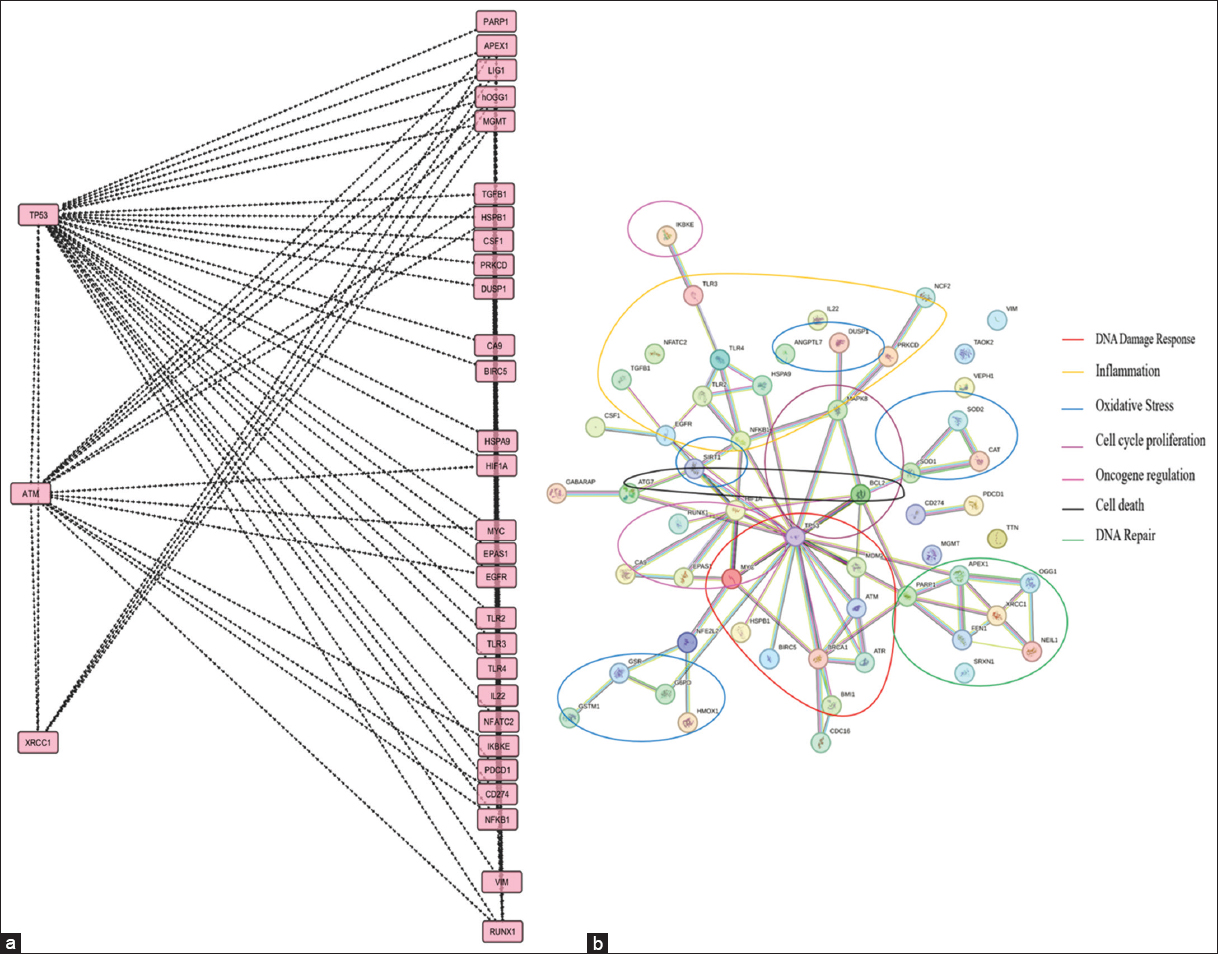
Breast cancer is the most common type of cancer in women diagnosed as of 2020, and it is the leading cause of cancer morbidity, disability, and mortality in women globally [14]. The RT remains a standard palliative therapy for curing breast cancer. The types of gene interactions may be categorized into gene regulatory networks, protein-protein interactions, regulation of gene expression, and crosstalk among pathways [15-17]. The coordinated cellular response to radiation depends on these gene-gene interactions, which affect the cancer patient’s response to RT. The genes represented in Figure 1 are known to be reported in previous literature, among which DNA repair and oxidative stress pathways are highly involved in RIAR. Also, the GSEA results comprising the genes and highly enriched pathways such as DNA repair and oxidative stress [Figure 2a-c] were in correlation with the gene-gene interaction results from Cytoscape (v 3.10.2) [Figure 3a] and STRING databases [Figure 3b]. p53 plays a main role in regulating other genes and is a major contributor for the occurrence of RIAR via seven major pathways [Figure 3b].
miRNAs serve as prognostic biomarkers for radio-resistance as reported in the literature [18]. miRNAs in this study can more effectively serve as potential targets for targeting genes involved in RIAR-associated mechanisms to enhance the effectiveness of RT. They are vital in controlling gene expression to enable the cells to respond to stress and cellular damage induced by radiation. Thus, miRNAs make cells adapt to radiation exposure, thereby promoting cell survival and maintaining genomic integrity; these correspond to the radio-adaptive response phenomenon. These can target genes in the homologous recombination and non-homologous end-joining pathway, thereby repairing the radiation-induced damages efficiently. miRNAs regulate genes involved in the oxidative stress response pathways, such as superoxide dismutase (SOD) and nuclear factor erythroid 2-related factor 2 such that cells have increased production of reactive oxygen species during radiation exposure [19].
Enrichr is a web-based enrichment analysis tool, which is simple to use and intuitive, providing an assortment of visualization summaries of the collective purposes of gene lists. Another noteworthy feature of the online analysis tool Enrichr is its comprehensiveness, ease of use, and interactive visualization of results. Enrichr tool predicts disease-specific signatures or transcriptional signatures. The tools used in this study identify disease-specific signatures via a comparison of gene expression patterns between the normal and diseased tissues [20]. Studies have reported the alterations in genes in various other cancers, i.e., about 381 genes have been associated with RT response in rectal cancers [21]. Genes such as SELP, PIM2, CCL19, SDS, NRP1, and SF3A2 were associated with RT sensitivity in cervical cancers [22].
miRNAs are long non-coding RNAs that target genes and have regulatory functions. miRNA microarray analysis in AG01522 normal human fibroblasts upon exposure to 5 cGy followed by 2 Gy has revealed the trigger of miRNAs upon low-dose-induced RIAR. Earlier in silico and in vitro studies have analyzed miRNAs as potential targets in atherosclerosis [23]. Two separate miRNA signatures comprising 4 up-regulated and 5 down-regulated miRNAs in breast tumors were identified from 1165 breast cancer patients retrieved from the Cancer Genome Atlas (TCGA-BRCA) [24]. miRNAs have oncogenic or tumour-suppressive functions and hence remain dysregulated in several cancers [25]. Alterations in the circulating miRNAs in blood plasma or serum can reflect the patient’s response to RT.
The miTarBase and miRNet enhance data usability and accessibility; thus, these are regarded as authentic databases for miRNA information. The enrichMiR tool seems to be highly advanced for interpretation and visualization of enrichment results due to its routine updates, also it uses the input data via integration with high-confidence and authentic databases like ScanMiR, TargetScan, miRTarBase & oRNAment. The enrichMiR tool provides results based on various choices such as overlap features of miRNA, and it also performs a comprehensive prediction based on miRNAs functionally relevant to target genes apart from statistically significant miRNAs [26]. The role of miRNAs associated with those 69 genes and breast cancer are discussed in Table 1. A network of miRNAs, namely miR-21, miR-155, and miR-205, has been identified as potential biomarkers for monitoring the radiation response in cancer patients [27-29]. miR-124 contributes to RIAR by either enhancing or suppressing inflammation and immune responses [27]. miR-17 downregulates p21 expression, leading to radio-resistance of cancer cells, thereby facilitating cell survival and proliferation [30]. miR-335 is involved in the DNA damage response pathway [31]. Overexpression of miR-106a leads to increased cell proliferation, conferring radio-resistance, and miR-128 can either enhance or suppress DNA damage response genes, leading to RIAR [32]. miR-185 enhances radiation-induced apoptosis and inhibits cell proliferation via targeting ATM and ATR [33]. miR-15a regulates the SMPD1, which is involved in apoptosis and inflammation upon radiation response [34]. miR-20a-5p targets Rab27b involved in apoptosis, altering radio-resistance [35]. miR-218 targets stress response pathways involved in radiation-induced stress [36]. miR-27a targets genes involved in DNA repair and cell survival [37]. miR-34a downregulates factors involved in cancer stem cell maintenance and thereby reduces cell survival, proliferation, and alters RIAR [38]. miR-93 serves to be a promising therapeutic target for enhancing RT outcomes in cancer treatment by increasing tumor radiosensitivity [39]. hsa-let-7a-5p regulates the balance between DNA repair-mediated radio-resistance and apoptosis-mediated radiosensitivity following radiation exposure [40]. miR-145 contributes to enhanced tumor radio-resistance; hence, targeting this can be more beneficial towards RT outcomes. miR-181a targets ATM and RAD52; hence, altering miR-181a can enhance DNA damage thereby reducing radio-resistance [41]. miR-24 enhances radiosensitivity by enhancing apoptosis and inhibiting cell proliferation [19]. miR-9-5p targets Suppressor of Cytokine Signaling 5, thereby exploring miR-9-5p can alter angiogenesis and cell survival, thereby altering RIAR [42]. miR-92a has been found to be involved in oxidative stress influencing RIAR by producing reactive oxygen species [43]. Hence, we found that about 19 miRNAs were found to be enriched in our study; these were majorly involved in the regulation of pathways such as oxidative stress, inflammation, DNA damage response, DNA repair, apoptosis, cell proliferation, and cell survival. Therefore, these 19 miRNAs can serve as predictive therapeutic targets to be explored more via further molecular approaches for the occurrence of RIAR in breast cancer. Exploring and monitoring the miRNAs in this study, i.e., the miRNAs associated with the RIAR phenomenon and those important in breast cancer may help in the dynamic assessment of RT efficacy in breast cancer patients. This can maximize the therapeutic efficacy in spite of the non-targeted effects of radiation.
miRNAs that regulate genes upon radiation exposure (also known as radio-miRs) act as gatekeepers in regulating the cellular mechanisms and pathways that lead to radio-resistance or RIAR [44]. This study has witnessed the involvement of about 19 miRNAs that modify several radio-adaptive response pathways notably, DNA repair, DNA damage response, cell survival, proliferation, and apoptosis via targeting RIAR genes and carcinoma of the breast. The tools used in this study remain advantageous due to the advancement in software/databases. A deeper understanding of these mechanisms and targeting miRNAs via molecules involved in these mechanisms can lead to more precise combination treatments. This is an in silico study based on the genes and miRNA targets available from databases. However, the targets mentioned in this study have to be evaluated using in vitro studies, either in blood samples of cancer patients or in cancer cell lines for analyzing and confirmation of targets being involved in RIAR during RT. The targets can be validated in breast cancer cell lines in in vitro by methods such as qRT-PCR, northern blotting, microarray analysis, next-generation sequencing, luciferase reporter assay, in situ hybridization, and specifically, western blot and flow cytometry for analysing the expression of proteins targeted by miRNAs. Combining these methods will give an understanding of miRNA roles in RIAR during RT, or therapy resistance. Evaluating in vitro will further confirm the effectiveness of the identified targets. Hence, this study serves as a preliminary basis for more futuristic lab studies for improving therapeutic efficacy.
REFERENCES
1. Jokhadze N, Das A, Dizon DS. Global cancer statistics:A healthy population relies on population health. CA Cancer J Clin. 2024;74:224-6. [CrossRef]
2. Wu Y, Song Y, Wang R, Wang T. Molecular mechanisms of tumor resistance to radiotherapy. Mol Cancer. 2023;22:96. [CrossRef]
3. Ananthanarayanan A, Raavi V, Srinivas Kondaveeti S, Ramachandran I, Perumal V. Insights on the radiation-induced adaptive response at the cellular level and its implications in cancer therapy. Cytogenet Genome Res. 2023;163:257-73. [CrossRef]
4. Gandhi NM. Cellular adaptive response and regulation of HIF after low dose gamma-radiation exposure. Int J Radiat Biol. 2018;94:809-14. [CrossRef]
5. Wiencke JK. Nicotinamide deficiency in human lymphocytes prevents the [3H]thymidine-induced adaptive response for the repair of X-ray-induced chromosomal damage. Exp Cell Res. 1987;171:518-23. [CrossRef]
6. Binkley MS, Diehn M, Eke I, Willers H. Mechanisms and Markers of Clinical Radioresistance. In:Willers H, Eke I, editors. Molecular Targeted Radiosensitizers, Cham:Springer International Publishing;2020. 63-96. [CrossRef]
7. Hou J, Wang F, Kong P, Yu PK, Wang H, Han W. Gene profiling characteristics of radioadaptive response in AG01522 normal human fibroblasts. PLoS One. 2015;10:0123316. [CrossRef]
8. Li G, Yang M, Ran L, Jin F. Expression profile of radiotherapy sensitive genes and tumor-associated immune cell infiltration and prognosis in multiple human cancers. J Radiat Res Appl Sci. 2022;15:5-11. [CrossRef]
9. Mohammadi C, Gholamzadeh Khoei S, Fayazi N, Mohammadi Y,Najafi R. RNA as promising theragnostic biomarkers for predicting radioresistance in cancer:A systematic review and meta-analysis. Crit Rev Oncol Hematol. 2021;157:103183. [CrossRef]
10. Joshi SC, Khan FA, Pant I, Shukla A. Role of radiotherapy in early breast cancer:An overview. Int J Health Sci (Qassim). 2007;1:259-64.
11. Bozhilova LV, Whitmore AV, Wray J, Reinert G, Deane CM. Measuring rank robustness in scored protein interaction networks. BMC Bioinformatics. 2019;20:446. [CrossRef]
12. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:05005. [CrossRef]
13. Haynes W. Benjamini-hochberg method. In:Encyclopedia of Systems Biology. New York:Springer;2013. 78-78. [CrossRef]
14. Mehrotra R, Yadav K. Breast cancer in India:Present scenario and the challenges ahead. World J Clin Oncol. 2022;13:209-18. [CrossRef]
15. Gollapalli P, Radhakrishna V, Kumari NS, Gnanasekaran TS. Elucidating genes and transcription factors of human peripheral blood lymphocytes involved in the cellular response upon exposure to ionizing radiation for biodosimetry and triage:An in silico approach. J Health Allied Sci NU. 2024;14:S35-50. [CrossRef]
16. Henkel L, Rauscher B, Boutros M. Context-dependent genetic interactions in cancer. Curr Opin Genet Dev. 2019;54:73-82. [CrossRef]
17. Sekaran TS, Kedilaya VR, Kumari SN, Shetty P, Gollapalli P. Exploring the differentially expressed genes in human lymphocytes upon response to ionizing radiation:A network biology approach. Radiat Oncol J. 2021;39:48-60. [CrossRef]
18. Al-Hawary SI, Abdalkareem Jasim S, Altalbawy FM, Kumar A, Kaur H, Pramanik A, et al. RNAs in radiotherapy resistance of cancer;a comprehensive review. Cell Biochem Biophys. 2024;82:1665-79. [CrossRef]
19. Kura B, Pavelkova P, Kalocayova B, Pobijakova M, Slezak J. MicroRNAs as regulators of radiation-induced oxidative stress. Curr Issues Mol Biol. 2024;46:7097-113. [CrossRef]
20. Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr:Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. [CrossRef]
21. Zhu M, Li X, Wang S, Guo W, Li X. Characterization of radiotherapy sensitivity genes by comparative gene set enrichment analysis. In:Huang DS, Jo KH, Zhang XL, editors. Intelligent Computing Theories and Application. Vol. 10955. Cham:Springer International Publishing;2018. 205-16. [CrossRef]
22. Wang Y, Ouyang Y, Cao X, Cai Q. Identifying hub genes for chemo-radiotherapy sensitivity in cervical cancer:A bi-dataset in silico analysis. Discov Onc. 2024;15:434. [CrossRef]
23. Mahjoubin-Tehran M, Aghaee-Bakhtiari SH, Sahebkar A, Butler AE, Oskuee RK, Jalili A. In silico and in vitro analysis of microRNAs with therapeutic potential in atherosclerosis. Sci Rep. 2022;12:20334. [CrossRef]
24. Samara M, Thodou E, Patoulioti M, Poultsidi A, Thomopoulou GE, Giakountis A. Integrated miRNA signatures:Advancing breast cancer diagnosis and prognosis. Biomolecules. 2024;14:1352. [CrossRef]
25. Kim T, Croce CM. MicroRNA:Trends in clinical trials of cancer diagnosis and therapy strategies. Exp Mol Med. 2023;55:1314-21. [CrossRef]
26. Soutschek M, Germade T, Germain PL, Schratt G. MiR predicts functionally relevant microRNAs based on target collections. Nucleic Acids Res. 2022;50:W280-9. [CrossRef]
27. Sheedy FJ. Turning 21: Induction of miR-21 as a key switch in the inflammatory response. Front Immunol. 2015;6:19. [CrossRef]
28. Robertson ED, Wasylyk C, Ye T, Jung AC, Wasylyk B. The oncogenic MicroRNA Hsa-miR-155-5p targets the transcription factor ELK3 and links it to the hypoxia response. PLoS One. 2014;9:113050. [CrossRef]
29. Cai J, Fang L, Huang Y, Li R, Yuan J, Yang Y, et al. R-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73:5402-15. [CrossRef]
30. Xu Z, Zhang Y, Ding J, Hu W, Tan C, Wang M, et al. R-17-3p downregulates mitochondrial antioxidant enzymes and enhances the radiosensitivity of prostate cancer cells. Mol Ther Nucleic Acids. 2018;13:64-77. [CrossRef]
31. Ye L, Wang F, Wu H, Yang H, Yang Y, Ma Y, et al. Functions and targets of miR-335 in cancer. Onco Targets Ther. 2021;14:3335-49. [CrossRef]
32. Pan YJ, Zhuang Y, Zheng JN, Pei DS. MiR-106a:Promising biomarker for cancer. Bioorg Med Chem Lett. 2016;26:5373-7. [CrossRef]
33. Wang J, He J, Su F, Ding N, Hu W, Yao B, et al. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013;4:699. [CrossRef]
34. Rana S, Espinosa-Diez C, Ruhl R, Chatterjee N, Hudson C, Fraile-Bethencourt E, et al. Differential regulation of microRNA-15a by radiation affects angiogenesis and tumor growth via modulation of acid sphingomyelinase. Sci Rep. 2020;10:5581. [CrossRef]
35. Huang D, Bian G, Pan Y, Han X, Sun Y, Wang Y, et al. MiR-20a-5p promotes radio-resistance by targeting Rab27B in nasopharyngeal cancer cells. Cancer Cell Int. 2017;17:32. [CrossRef]
36. Schell G, Roy B, Prall K, Dwivedi Y. R-218:A stress-responsive epigenetic modifier. Noncoding RNA. 2022;8:55. [CrossRef]
37. Cho WC. MicroRNAs in cancer - from research to therapy. Biochim Biophys Acta (BBA) Rev Cancer. 2010;1805:209-17. [CrossRef]
38. Li WJ, Wang Y, Liu R, Kasinski AL, Shen H, Slack FJ, et al. MicroRNA-34a:Potent tumor suppressor, cancer stem cell inhibitor, and potential anticancer therapeutic. Front Cell Dev Biol. 2021;9:640587. [CrossRef]
39. Chen X, Liu J, Zhang Q, Liu B, Cheng Y, Zhang Y, et al. Exosome-mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J Exp Clin Cancer Res. 2020;39:65. [CrossRef]
40. Unger K. Integrative characterisation and prediction of the radiation response in radiation oncology. In:Cumulative Habilitation Thesis for Obtaining the Venia Legendi in the Field of Radiation Biology;2018.
41. Bisso A, Faleschini M, Zampa F, Capaci V, De Santa J, Santarpia L, et al. Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle. 2013;12:1679-87. [CrossRef]
42. Wei YQ, Jiao XL, Zhang SY, Xu Y, Li S, Kong BH. MiR-9-5p could promote angiogenesis and radiosensitivity in cervical cancer by targeting SOCS5. Eur Rev Med Pharmacol Sci. 2019;23:7314-26.
43. Jiang M, Li X, Quan X, Li X, Zhou B. MiR-92a family:A novel diagnostic biomarker and potential therapeutic target in human cancers. Front Mol Biosci. 2019;6:98. [CrossRef]
44. Pedroza-Torres A, Romero-Córdoba SL, Montaño S, Peralta-Zaragoza O, Vélez-Uriza DE, Arriaga-Canon C, et al. Radio-miRs:A comprehensive view of radioresistance-related microRNAs. Genetics. 2024;227:iya097. [CrossRef]