1. INTRODUCTION
Pathogenic microbes are responsible to cause various infectious illnesses. It can be transmitted either directly or indirectly among individuals through animal and insect carriers or through ingestion of contaminated food, water, and various environmental factors [1]. Lately, there has been a rise in resistance among conventional antibiotics by both Gram-negative bacteria and Gram-positive bacteria [2]. Antibiotic-resistant bacteria have rapidly emerged as a major global health concern, rendering many traditional antibiotics useless and necessitating the urgent development of alternative antimicrobial methods. In this context, Silver nanoparticles (AgNPs) have been reported as the most effective bactericide and bacteriostatic agent against both Gram-positive and Gram-negative bacteria, including multidrug-resistant strains. Unlike conventional antibiotics, which usually target specific bacterial pathways, AgNPs have various modes of action, making it harder for bacteria to build resistance. These mechanisms include chemical production of reactive oxygen species (ROS), physical interactions with the bacterial membrane, and biochemical interference with essential cellular functions such as protein synthesis and DNA replication, leading to cell death. Multiple functionality of AgNPs not only increases their bactericidal efficacy but also consider them as a viable substitute in the battle against antibiotic resistance [3].
However, different chemical and physical approaches have been employed to synthesize AgNPs. However, these approaches produces hazardous chemical residues, high energy consumption, and environmental risks which reduces the sustainability and biocompatibility of AgNPs. In order to address these obstacles, scientists are increasingly using green synthesis techniques, which use biological systems sch as fungi, bacteria, and plant extracts to create AgNPs in a way that is safe for the environment, economical, and biological. In this method, biological entities produce natural capping and stabilizing compounds [4]. It has been demonstrated that tollens, polysaccharides polyoxometalate, polyphenols, and other secondary metabolites extracted from biological entities are effective in reducing the harmful chemical substances. According to reports, bio-surfactants and extracts produced by bacteria, yeast, fungi, or plants are utilized as capping and reducing agents in biological reduction. These are the alternatives to reducing and capping agents utilized in the chemical synthesis process. This technique enhances the therapeutic potential of AgNPs while simultaneously adhering to green chemistry principles [5]. Furthermore, in the bacterial synthesis process of AgNPs, bacterial metabolites such as enzymes, organic acids, amino acids, and sugars play an important role in nanoparticle production by acting as reducing agents. They convert metal ions (e.g., Ag+) to zero-valent metal forms (e.g., Ag0). Bacterial cell membranes contain metabolites such as proteins, polysaccharides, lipids, and other biopolymers that can adhere to the surface of newly produced nanoparticles, capping them and preventing aggregation. This is a critical step in nanoparticle production. This biologically driven approach not only provides fine control over nanoparticle size and form, but it also improves biocompatibility, making bacterial-mediated synthesis a promising alternative to traditional chemical procedures [6].
In a study, Prabhu and Poulose, in 2012 [7] have been reported that initially silver nitrate was synthesized from bacterial strain Pseudomonas stutzeri AG259, identified from a silver mine. Moreover, it has been reported that 50 nm particles sized AgNPs were fabricated by the bacterial species Bacillus licheniformis, identified from collected waste of municipal sewage [8]. Several reports have shown that AgNPs exhibit some remarkable properties, such as antibacterial, antioxidant, optical properties, large surface area to volume ratio, and higher efficacy. Due to which AgNPs have been found effective against most of the pathogens such as Salmonella paratyphi Pseudomonas aeruginosa, Bacillus subtilis, Escherichia coli Stapylococcus aureus, and Klebsiella pneumoniae was reported by Mohammed et al., [9], Bindhu et al., [10]; Prasad and Elumalai [11].
In the present work, the extracellular biosynthesis method is being studied with Enterobacter mori C29(1)CG, which was isolated from contaminated soil. X-ray diffraction (XRD), Fourier transform infrared (FTIR), scanning electron microscopy (SEM), and ultraviolet (UV)-visible spectrophotometry were used to characterize the produced AgNPs. Therefore, the goal of this work is to assess the antibacterial activity (in vitro) of these green-generated nanoparticles against human pathogens using well-diffusion method. The study illustrated that AgNPs have strong antibacterial properties and have the potential to develop into a brand-new class of antimicrobial drugs for the treatment of bacterial infections in humans.
2. MATERIALS AND METHODS
2.1. Isolation of the Bacteria
Eight bacterial strains were isolated from a contaminated site in the Gwalior region, India. Serially diluted soil samples were prepared up to 106 dilution. About 100 μL of each dilution was placed on Nutrient Agar medium and kept in an incubator under aseptic conditions at 37ºC for 24 h. All isolated strains were selected for further studies.
2.2. Bacterial Synthesis of AgNPs
The isolates were screened to obtain an efficient strain that synthesized AgNPs. All bacterial strains were cultivated in LB (Luria-Bertani) broth medium at 37°C for 48 h on an incubator shaker at 180 rpm. After incubation, the culture supernatant was collected by centrifugation at 10,000 rpm for 8 min. AgNPs were produced by treating an aqueous solution of 10 mM AgNO3 with culture supernatant [Figure 1] at 200 rpm for 48 h at 30°C. As a control, no AgNO3 was added to the supernatant [12]. The culture supernatant was centrifuged at 10,000 rpm for 8 min, and the pellet was thoroughly washed 3 times with sterile distilled water to eliminate any medium residues. The cell-free supernatant of the bacterial strain was obtained by filtration via Whatman filter paper No. 1. The cell-free bacterial supernatants were utilized for the synthesis of AgNPs [13]. Positive synthesis of AgNPs was determined based on the color change to brown [12]. On the basis of rapid color change, an efficient strain was further selected for molecular sequencing.
 | Figure 1: Schematic representation of the synthesis of silver nanoparticles. [Click here to view] |
2.3. Molecular Sequencing of the Selected Bacterial Isolates
The selected isolate was identified by analyzing 16s ribosomal RNA (rRNA) sequences. The HiPurA Bacterial DNA Purification Spin Column Kit (MB505-250PR, HiMedia, India) was used to isolate genomic DNA, which was then examined using 1% agarose gel electrophoresis. Primers F27 (5’AGAGTTTGATCMTGGCTCAG3’) and 1492R (5’ GGTTACCTTGTT ACGACTT 3’) were used to amplify the bacterial-specific 16s rRNA gene (1500 bp) [14]. Polymerase chain reaction (PCR) amplification was performed utilizing an Applied Biosystems Veriti Thermal Cycler with the following parameters: denaturation at 94°C for 5 min, succeeded by 34 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min, concluding with a final extension at 72°C for 7 min. PCR results were sequenced by NCIM CSIR-NCL in Pune. The DNA sequence was submitted to GenBank for homology analysis using the BLASTN tool [15]. The DNA sequence was submitted to NCBI via GenBank, and multiple sequence alignment was conducted using Clustal W [16]. The phylogenetic tree was built using the neighbor-joining method in MEGA 11 software [17], with a 1000-iteration bootstrap [18] and evolutionary relationships were estimated.
2.4. Characterization of AgNP
2.4.1. UV-vis spectra analysis
The reduction of pure silver ions was confirmed by analyzing the UV-vis spectra of the solution at ambient temperature using a UV-visible spectrophotometer (UV 1800 Shimadzu) throughout the wavelength range of 350–550 nm. The maximum absorption wavelength would be achieved by surface plasmon resonance [19].
2.4.2. Powder XRD
An XRD, Rikagu Miniflex 600 with Cu-Kα radiation (k = 0.1541 nm) was used for recording the XRD pattern. Data were captured between 10° and 80° (2θ) at a scan speed of 4 min−1. The XRD technique was utilized to ascertain the particle size employing Scherrer’s formula:
D = kλ/β.Cosθmax
Where d represents the average crystal size, k denotes the X-ray wavelength (0.1541 nm), b signifies the full-width at half-maximum, and h indicates the diffraction angle [19].
2.4.3. FTIR
A Perkin-Elmer, Spectrum Two, FTIR spectrometer was used to characterize the functional groups on the surface of biosynthesized AgNPs. The range of 4000–500/cm was used for recording the FTIR spectrum [20].
2.4.4. SEM and EDX analysis of AgNPs
SEM (SEM, Philips, Model: Quanta 200 FEG) was used to analyze the morphology and structure of the sample. In addition, the elemental composition of the produced AgNPs was determined using an energy dispersive X (EDX) ray spectrometer [21].
2.5. Antimicrobial Activity
The agar well-diffusion method was conducted to evaluate the antibacterial efficacy of the biosynthesized AgNPs according to Salari et al., with slight modification [19]. The in vitro antibacterial activity of AgNPs was assessed against the following three test pathogenic strains. S. aureus (MTCC 740), Klebsiella pneumonia (MTCC 109), and Streptococcus mutans (MTCC 890). S. aureus and K. pneumonia were inoculated in nutrient broth for 24 h at 37ºC and S. mutans was grown in Enriched brain heart infusion broth for 24 h at 37ºC. A sterilized cork borer was employed to form wells with a diameter of 6 mm on Muller–Hinton agar medium. A cotton swab saturated with each test strain was employed to uniformly inoculate the agar medium. Each well on the plates received three separate additions of approximately 100 μL of nanosolution at four distinct concentrations: 100µg/mL, 75µg/mL, 50µg/mL, and 25µg/mL. The assay included positive and negative controls. The antibiotic was used as a positive control (PC), with distilled water serving as a negative control. Plates were held upright at 37°C for 24 h to observe the zone of inhibition. Using a zone reader scale (Himedia Pvt. Ltd.), the diameter of the inhibition zones (mm) surrounding the wells was measured to assess the antibacterial activity. The assay was performed in triplicate.
3. RESULT AND DISCUSSION
A total of 8 strains, namely, C11, C15, C28, C29(1)CG, C33, C40(3), C54, C63, were isolated from contaminated soil of the Gwalior region and used for nanoparticles synthesis. Based on the rapid reduction of Ag metal ions, C29(1)CG was chosen and identified for 16SrRNA molecular sequencing.
3.1. Molecular Identification of Selected Bacterial Strains
The selected isolated bacteria C29(1)CG were first observed under a light microscope and recorded as Gram-negative, short rods, and motile. The morphology of the bacterial colony was smooth, soft, and white. The phylogenetic position of the bacterial isolate C29(1)CG was determined by analyzing the 16S rRNA gene sequencing and comparing it to closely related E. mori species from the GenBank database. The isolated species was identified as E. mori C29(1)CG (accession no. PQ394970) with a percentage similarity of 99% as shown in Figure 2.
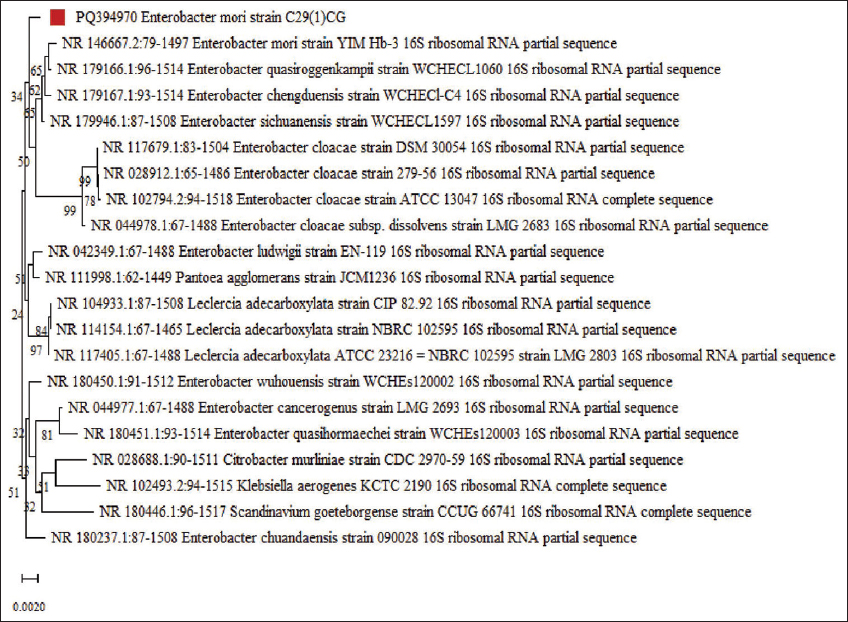 | Figure 2: Phylogenetic analysis of ribosomal RNA gene sequence of strain PQ394970 constructed by neighbor joining method with 1000 bootstraps (Genbank accession no. PQ394970). [Click here to view] |
In this study, AgNPs has been synthesized with cell cell-free supernatant of E. mori C-29(1)CG, which is isolated from contaminated soil. Prior research has mostly concentrated on species such as Enterobacter cloacae for the production of AgNPs, owing to its extensively documented metabolic adaptability and capacity to reduce metal ions [22,23].
It has been demonstrated that E. cloacae secretes metabolites and reductive enzymes that aid in the stability of nanoparticles and the reduction of Ag ions. E. mori, on the other hand, is a species that has received relatively little attention in nanobiotechnology. Despite its phylogenetic resemblance to E. cloacae and capacity to generate extracellular bio-reductants, little is known about its function in nanoparticle synthesis to date. By providing a novel microbial contender that may produce unique nanoparticle morphologies, stability profiles, or bioactivities, this work pioneers the use of E. mori in the environmentally friendly synthesis of AgNPs.
3.2. Characterization of AgNPs by UV-Vis Spectrophotometer
AgNPs synthesized by the cell-free supernatant of C29(1)CG was characterized by UV-Vis spectrophotometer, that conducted the primary assessment of nanoparticles. The color of the AgNP mixture that progressively changed from white to yellowish-brown indicates the formation of biosynthesized AgNPs was then used for spectral analysis. The result revealed that the highest peak for biosynthesized AgNPs was recorded at 410 nm [Figure 3]. This peak is characteristic of AgNPs synthesis and is caused by the excitation of surface plasmon vibrations in the nanoparticles [24]. In contrast, prior research suggested that AgNPs possessed free electrons, creating a surface plasmon resonance at 406, 416, 430, and 448 nm [25-29].
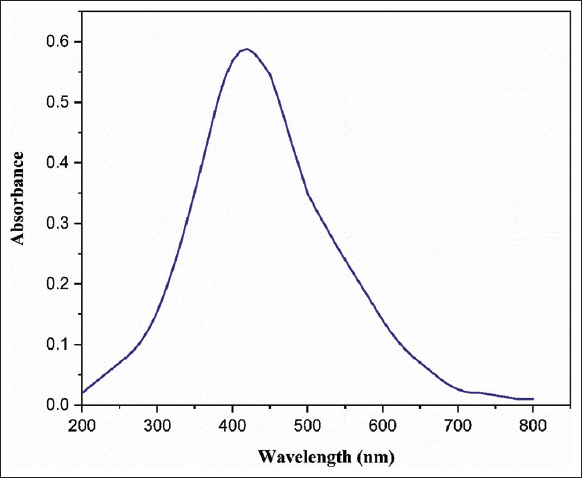 | Figure 3: Ultraviolet-vis absorption spectra of silver nanoparticles synthesized using cell-free supernatant of Enterobacter mori C-29(1)CG. [Click here to view] |
3.3. Characterization of AgNPs by XRD
This method is extensively employed to examine the crystalline structure of AgNPs. The XRD analysis facilitated the identification and quantification of the unique crystallographic structure of this substance present in the bacterial extract, together with the particle sizes of the produced AgNPs. The crystalline nature of the biosynthesized AgNPs was shown by their XRD spectra, and the particles’ oxidation state was ascertained over time. The findings showed that AgNPs were present on the nanometric scale. The Debye-Scherrer formula was used to get the average particle size [30]. The dry sample of biosynthesized AgNPs was further characterized by XRD, which confirms its crystalline nature. The XRD analysis represents the Miller-Bravais indices of (111), (200), (142), (220), and (311) as shown in Figure 4. This reveals that the structure of AgNPs is a face-centered cubic structure as per available literature (JCPDS, File No. 4-0783). Similar results were achieved by Fouad et al., and Giri et al. [12,31]. The crystallite sizes of silver in various planes were measured at 34.20 nm, 34.92 nm, and 30.27 nm, with a mean value of 33.13 nm across all peaks.
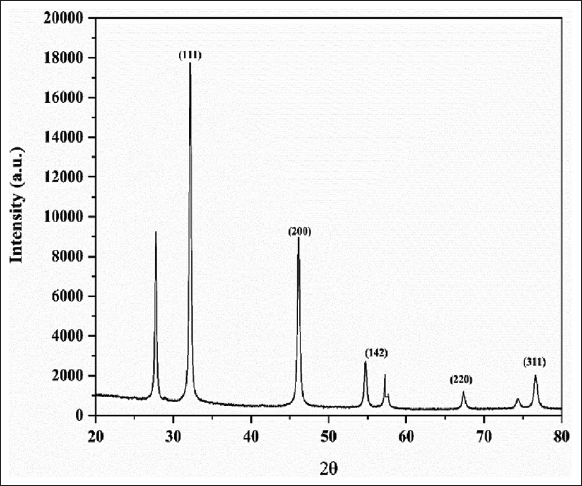 | Figure 4: X-ray diffraction pattern of silver nanoparticles synthesized from cell-free supernatant of Enterobacter mori C-29(1)CG. [Click here to view] |
3.4. Characterization of AgNPs by FTIR
FTIR Spectroscopy is an essential analytical technique in investigating the role of bacterial extract in the reduction and stabilization of AgNPs. In this method, infrared light is utilized to assess the surface chemistry of nanoparticles fabricated by bacterial extract [30]. Infrared spectrum studies were carried out to confirm the functional groups and chemical bonds of the phytochemicals present in the extract using the absorption and transmittance values of infrared radiation. The standard spectra of AgNPs employing cell-free supernatant from C-29(1)CG were shown. The peaks were shown at 2918, 1635, 1526, 1386, 1230, 1049, 530.7 cm−1. Asymmetric stretching of a methyl group - CH3 and C-H stretching of alkanes or secondary amine are responsible for the peak at ~2918 cm-−1. The peak at ~1633.91 cm−1 and 1526 cm−1 is typically associated with amide I (C = O stretching of proteins). Moreover, peak at 1386 and 1230 cm−1 represents the presence of C-N aromatic amino group. Singh and Mijakovic [32] achieved similar outcomes. The (C-O) of an alkoxy group may be responsible for the peaks at 1049 and 530.7 cm−1 indicate C-C deformation as shown in Figure 5 [32,33]. The result of FTIR represents the presence of carboxyl groups (−C = O) and amine groups (NH), which indicate the presence of proteins, amino acids, and other biomolecules extracted from the supernatant of the E. mori C-29(1)CG. These protein molecules act as reducing, capping, and stabilizing agents and are responsible for reducing Ag+ to Ag°0 and attaching to the surface of AgNPs by –NH and C = O groups and preventing their agglomeration [34]. These compounds may enhance hydrogen bonding or electrostatic contact with bacterial membranes, enhancing AgNP adherence and penetration into microbial cells. Furthermore, they regulate redox reactions at the interface between nanoparticles, which increases the production of ROS [35]. Therefore, FTIR spectroscopy is an easy and suitable way to ascertain the role of bacterial extracts in the reduction of AgNPs.
 | Figure 5: Fourier transform infrared spectroscopy of silver nanoparticles synthesized using cell-free supernatant of Enterobacter mori C-29(1)CG. [Click here to view] |
3.5. Morphological Analysis of AgNPs
Field emission SEM was used to examine the shape and particle size of the sample. The freeze-dried biosynthesized AgNPs were measured under the FESEM at 3kV and are clearly displayed in Figures 6 and 7. The structure of most of these AgNPs resembles an irregular spherical that is uniformly distributed. In addition, Fouad et al. [12] demonstrated that AgNPs generated with B. subtilis and Bacillus amyloliquefaciens have similar morphological features. Furthermore, the EDX instrument verified that the silver element was present in the AgNPs. This shows that the AgNPs were formed in the bacterial cell-free supernatant of C29(1)CG. The reduction of silver ions Ag+ to Ag0 was confirmed by the prominent peak of silver ions recorded at 3 KeV [Figure 7]. Fouad et al., reported the same peak (3 keV) [12]. The benefit of SEM combined with an EDX-ray spectrometer provides a powerful characterization tool by offering both high-resolution images of surface topography and elemental analysis.
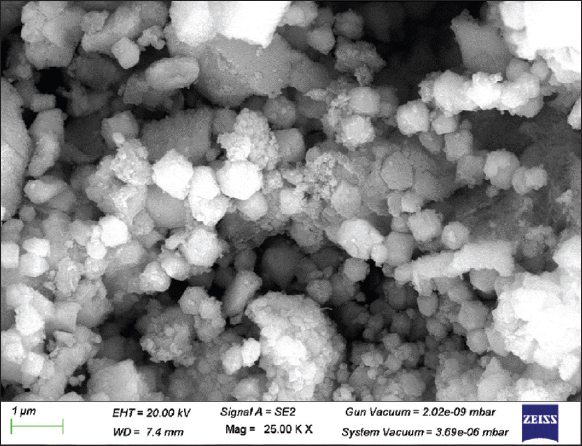 | Figure 6: Scanning electron microscopy image of silver nanoparticles synthesized using cell-free supernatant of Enterobacter mori C-29(1)CG. [Click here to view] |
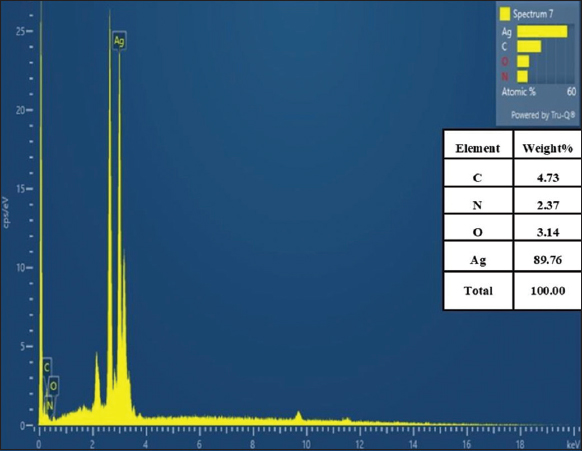 | Figure 7: Energy dispersive X-ray pattern of silver nanoparticles synthesized using cell-free supernatant of Enterobacter mori C-29(1)CG. [Click here to view] |
3.6. Antimicrobial Activity
The inhibitory effect of AgNPs was assessed against microorganisms. However, all four different concentrations of biosynthesized AgNPs against test microbes are shown in Table 1 and Figure 8. At a concentration of 100 μg/mL concentration, biosynthesized AgNPs exhibited a significant zone of growth inhibition against S. aureus, S. mutans, and K. pneumoniae in comparison to the control, as shown in Figure 9 and Table 1. Biosynthesized AgNPs showed the zone of inhibition size for S. aureus, S. mutans, and K. pneumoniae was 22.667 ± 0.882, 22.333 ± 0.333, and 12.667 ± 0.333, respectively. Furthermore, the study explains that 75, 50, and 25 μg/mL concentrations of biosynthesized AgNPs have not significantly enhanced the antibacterial activity as shown by 100 μg/mL concentrations against the test microorganisms [Figure 8 and Table 1]. Consequently, it is revealed that a concentration of 100 μg/mL of biosynthesized AgNPs may serve as an effective antibacterial agent against S. aureus followed by S. mutans and K. pneumoniae. Biosynthesized AgNPs was compared to Streptomycin (26.333 ± 0.333 for S. aureus, 25.667 ± 0.333 for S. mutans and 24.5 ± 0.408 for K. pneumoniae) as PC. However, distilled water as a negative control did not show any activity against test microorganisms. Hereby, in case of S. aureus and S. mutans, the present study reveals that the increasing concentration of AgNPs as antibacterial agent increases the zone of inhibition size and hence, increases the antibacterial activity against disease-causing microorganisms. In a similar study, Fouad et al., [12] stated that the increased concentration of AgNPs, gave the highest diameter of inhibition growth against test microorganism. AgNPs exhibits various mechanisms of antimicrobial action against resistant pathogens. The study reveals that biosynthesized AgNPs showed the strongest antibacterial efficacy against S. aureus among the studied pathogens. The Gram-positive cell wall structure of S. aureus has a thick peptidoglycan layer but no outer membrane, is responsible for this greater susceptibility [36]. n addition, S. aureus is well-known for having a large number of membrane-associated proteins, which could facilitate interactions with AgNPs that have been functionalized with proteins. Biosynthesized AgNPs are very efficient against this pathogen because of their ability to increase binding, cause ROS-mediated damage, and disrupt cells [37]. However, Guzman et al., [38] have been reported that the antimicrobial potential of AgNPs was mainly due to the close adhesion of nanoparticle’s surface to microbial cells. It is stated that during the antibacterial impact, silver ion extensively interact with phosphorus and sulfur. These elements are found in abundance across the cell membrane and silver ions exploit these key materials for showing antibacterial ability. Moreover, AgNPs also disrupt membranes and trigger the generation of ROS, producing free radicals with a potent bactericidal activity, ultimately causing microbial cell death [39]. Furthermore, AgNPs disrupt protein activity and damage DNA in bacterial cell. The most generally recorded bactericidal mechanism involves AgNPs interacting with the thiol groups of L-cysteine residues in proteins, resulting in their inactivation. This interaction can denature RNA and DNA by breaking the respiratory chains. The potential of AgNPs to perforate into bacterial cells promotes damage on morphology and structure and results into cell death [3].
Table 1: Antimicrobial effect of AgNPs synthesized with cell free supernatant of strain C-29 (1) CG against test pathogenic microorganisms (Zone of inhibition in mm)
| Sample name | Staphylococcus aureus | Streptococcus mutans | Klebsiella pneumoniae | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Concentrations in µg/ml | ||||||||||||
| 100 | 75 | 50 | 25 | 100 | 75 | 50 | 25 | 100 | 75 | 50 | 25 | |
| AgNPs synthesized with strain C-29 (1) | 22.667±0.882 | 18.333±0.333 | 9.667±0.333 | 0±0 | 22.333±0.333 | 11.667±0.333 | 0±0 | 0±0 | 12.667±0.333 | 0±0 | 0±0 | 0±0 |
| Positive control | 26.333±0.333 | 25.333±0.333 | 20.333±0.333 | 14.333±0.333 | 25.667±0.333 | 20±0.577 | 0±0 | 0±0 | 24.5±0.408 | 22.667±0.333 | 20.333±0.333 | 0±0 |
| Negative control | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Where, NA=No activity. Values represent the mean of three replicates±SD. Significant differences were found against all three pathogens in all four concentration. The probability values of **P<0.01, ***P<0.001 and ****P<0.0001 were compared to the control.
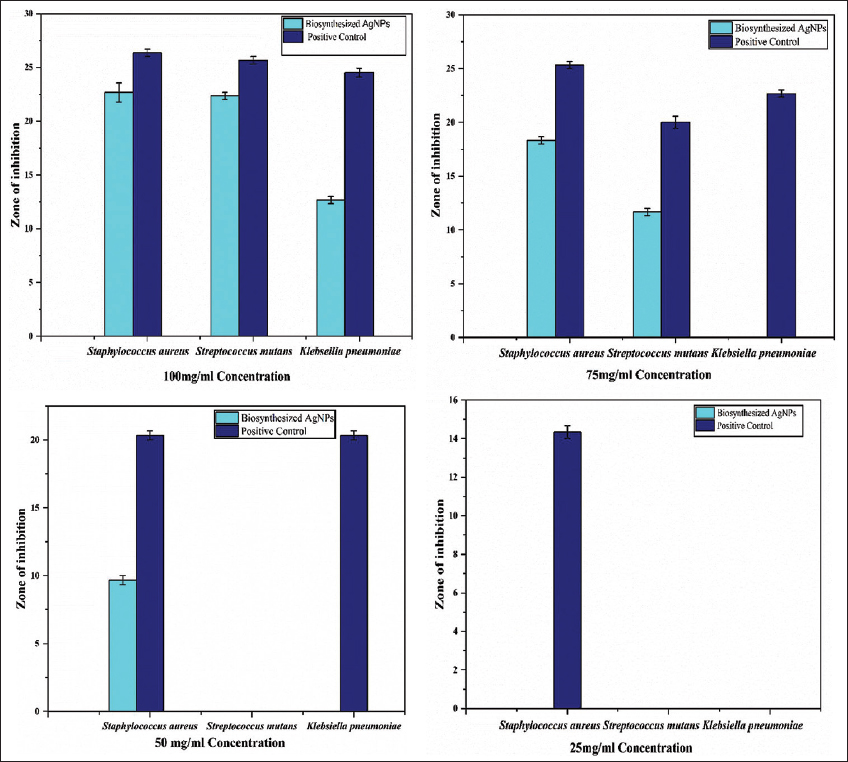 | Figure 8: Graphical representation of effect of silver nanoparticles synthesized from cell supernatant of Enterobacter mori C-29(1)CG with four different concentrations against Staphylococcus aureus, Streptococccus mutans and Klebsiella pneumonia as comparison to standard drug. Error bars in each column represent the standard error within same treatment. [Click here to view] |
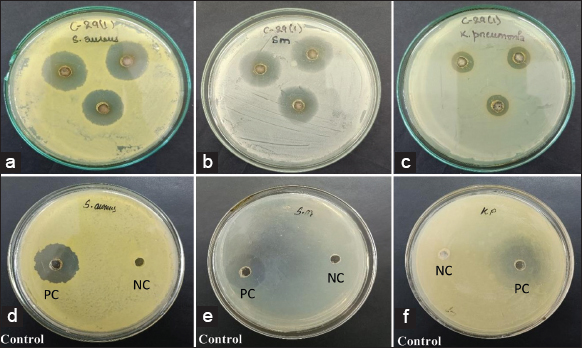 | Figure 9: Effect of AgNPs synthesized using cell-free supernatant of Enterobacter mori C-29(1)CG on (a) Staphylococcus aureus, (b) Streptococcus mutans and (c) Klebsiella pneumoniae showing clear zone of inhibition indicating strong antibacterial action while (d),(e) and (f) shows positive control (PC) and negative control (NC). [Click here to view] |
3.7. Statistical Analysis
Results are presented as mean ± standard error of the mean. All data were statistically analyzed using one-way analysis of variance (ANOVA) using GraphPad Prism version 3. The significance of the difference was determined using one-way ANOVA, followed by Dunnett’s multiple comparison test. Significant differences were found against all three pathogens in all four concentrations. The probability values of **P < 0.01, ***P < 0.001, and ****P < 0.0001 were compared to the control.
4. CONCLUSION
Green route of nanoparticles synthesis is one of the most intense fields of research in recent time. The study investigated the use of cell-free supernatant of E. mori C29(1)CG to synthesize AgNPs and its application in antimicrobial activity. However, isolated soil bacteria have shown a remarkable ability to convert silver-ion-charged to silver-ion-free under laboratory conditions. This technique could be convenient, nontoxic, and cost-effective for the production of AgNPs at a commercial level. Furthermore, these AgNPS were confirmed and have shown remarkable results by the analysis of UV-visible spectroscopy, XRD, FTIR, SEM, and EDX. The current finding revealed that biosynthesized AgNPs are considered more powerful antimicrobial agent against S. aureus followed by S. mutans and K. pneumoniae as shown in the figure. Hereby, biosynthesized AgNPs provide a promising strategy to combating antibiotic resistance by disrupting bacterial processes. Hence, the study benefits the pharmaceutical industry and provides an ideal basis for future research into the development of innovative medications. Moreover, this green technology includes future prospects in assessing cytotoxicity, performing in vivo testing, or scaling up production.
5. Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
6. Funding
There is no funding to report.
7. Conflicts of interest
The authors report no financial or any other conflicts of interest in this work.
8. Ethical approvals
This study does not involve experiments on animals or human subjects.
9. Data availability
All the data is available with the authors and shall be provided upon request.
10. Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
11. Use of artificial intelligence (AI)-assisted technology
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. National Institutes of Health. Biological Sciences Curriculum Study. NIH Curriculum Supplement Series. Information about Mental Illness and the Brain. Understanding Emerging and Re-emerging Infectious Diseases. Bethesda, MD:National Institutes of Health (US);2007.
2. Frieri M, Kumar K, Boutin A. Antibiotic resistance. J Infect Public Health. 2017;10(4):369-78. [CrossRef]
3. More PR, Pandit S, Filippis AD, Franci G, Mijakovic I, Galdiero M. Silver nanoparticles:Bactericidal and mechanistic approach against drug resistant pathogens. Microorganisms. 2023;11(2):369. [CrossRef]
4. Gupta C, Rabani MS, Gupta MK, Tripathi S, Pathak A. Nanoscience in biotechnology. In:Diverse Applications of Nanotechnology in the Biological Sciences. United States:Apple Academic Press;2022. 241-67. [CrossRef]
5. Gupta C, Gupta MK, Rabani MS, Tripathi S, Pathak A. Green synthesized nanomaterials as tools to remediate aquatic pollution. Aquat Contam Tolerance Bioremediation. 2024:277-89. [CrossRef]
6. Lahiri D, Nag M, Sheikh HI, Sarkar T, Edinur HA, Pati S, et al. Microbiologically-synthesized nanoparticles and their role in silencing the biofilm signaling cascade. Front Microbiol. 2021;12:636588. [CrossRef]
7. Prabhu S, Poulose EK. Silver nanoparticles:Mechanism of antimicrobial action, synthesis, medical applications, and toxicity effects. Int Nano Lett. 2012;2:32. [CrossRef]
8. Kalimuthu K, Babu RS, Venkataraman D, Bilal M, Gurunathan S. Biosynthesis of silver nanocrystals by Bacillus licheniformis. Colloids Surf B Biointerfaces. 2008;65(1):150-3. [CrossRef]
9. Mohammed AB, Mohamed A, El-Naggar NE, Mahrous H, Nasr GM, Abdella A, et al. Antioxidant and antibacterial activities of silver nanoparticles biosynthesized by Moringa oleifera through response surface methodology. J Nanomater. 2022;2022(1):9984308. [CrossRef]
10. Bindhu MR, Umadevi M, Esmail GA, Al-Dhabi NA, Arasu MV. Green synthesis and characterization of silver nanoparticles from Moringa oleifera flower and assessment of antimicrobial and sensing properties. J Photochem Photobiol B Biol. 2020;205:111836. [CrossRef]
11. Prasad TN, Elumalai EK. Biofabrication of Ag nanoparticles using Moringa oleifera leaf extract and their antimicrobial activity. Asian Pac J Trop Biomed. 2011;1(6):439-42. [CrossRef]
12. Fouad H, Hongjie L, Yanmei D, Baoting Y, El-Shakh A, Abbas G, et al. Synthesis and characterization of silver nanoparticles using Bacillus amyloliquefaciens and Bacillus subtilis to control filarial vector Culex pipiens pallens and its antimicrobial activity. Artif Cells Nanomed Biotechnol. 2017;45(7):1369-78. [CrossRef]
13. Soliman H, Elsayed A, Dyaa A. Antimicrobial activity of silver nanoparticles biosynthesised by Rhodotorula sp. Strain ATL72. Egypt J Basic Appl Sci. 2018;5(3):228-33. [CrossRef]
14. Clarridge JE 3rd. Impact of 16S rRNA gene sequence analysis for identification of Bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev. 2004;17(4):840-62. [CrossRef]
15. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403-10. [CrossRef]
16. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W:Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673-80. [CrossRef]
17. Tamura K, Stecher G, Kumar S. MEGA11:Molecular evolutionary genetics analysis version 11. Molecul Biol Evol. 2021;38(7):3022-7. [CrossRef]
18. Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution. 1985;39(4):783-91. [CrossRef]
19. Salari Z, Danafar F, Dabaghi S, Ataei SA. Sustainable synthesis of silver nanoparticles using macroalgae Spirogyra varians and analysis of their antibacterial activity. J Saudi Chem Soc. 2016;20(4):459-64. [CrossRef]
20. Buszewski B, Railean-Plugaru V, Pomastowski P, Rafi?ska K, Szultka-Mlynska M, Golinska P, et al. Antimicrobial activity of biosilver nanoparticles produced by a novel Streptacidiphilus durhamensis strain. J Microbiol immunol Infect. 2018;51(1):45-54. [CrossRef]
21. Urnukhsaikhan E, Bold BE, Gunbileg A, Sukhbaatar N, Mishig-Ochir T. Antibacterial activity and characteristics of silver nanoparticles biosynthesized from Carduus crispus. Sci Rep. 2021;11(1):21047. [CrossRef]
22. Shahverdi AR, Minaeian S, Shahverdi HR, Jamalifar H, Nohi AA. Rapid synthesis of silver nanoparticles using culture supernatants of Entero Bacteria:A novel biological approach. Process Biochem. 2007;42(5):919-23.[CrossRef]
23. Ashraf N, Ahmad F, Jing Jie C, Tuo Di Z, Feng-Zhu Z, Yin DC. Optimization of Enterobacter cloacae mediated synthesis of extracellular silver nanoparticles by response surface methodology and their characterization. Part Sci Technol. 2020;38(8):931-43. [CrossRef]
24. Shankar SS, Rai A, Ahmad A, Sastry M. Rapid synthesis of Au, Ag, and bimetallic Au core-Ag shell nanoparticles using neem (Azadirachta indica) leaf broth. J Colloid Interface Sci. 2004;275(2):496-502. [CrossRef]
25. Banu AN, Balasubramanian C, Moorthi PV. Biosynthesis of silver nanoparticles using Bacillus thuringiensis against dengue vector, Aedes aegypti (Diptera:Culicidae). Parasitol Res. 2014;113:311-6. [CrossRef]
26. Du J, Singh H, Yi TH. Biosynthesis of silver nanoparticles by Novosphingobium sp. THG-C3 and their antimicrobial potential. Artif Cells Nanomed Biotechnol. 2017;45(2):211-7. [CrossRef]
27. Gopinath V, Velusamy P. Extracellular biosynthesis of silver nanoparticles using Bacillus sp. GP-23 and evaluation of their antifungal activity towards Fusarium oxysporum. Spectrochim Acta A Mol Biomol Spectrosc. 2013;106:170-4. [CrossRef]
28. Sundaravadivelan C, Padmanabhan MN. Effect of mycosynthesized silver nanoparticles from filtrate of Trichoderma harzianum against larvae and pupa of dengue vector Aedes aegypti L. Environ Sci Pollut Res Int. 2014;21:4624-33. [CrossRef]
29. Wang C, Kim YJ, Singh P, Mathiyalagan R, Jin Y, Yang DC. Green synthesis of silver nanoparticles by Bacillus methylotrophicus, and their antimicrobial activity. Artif Cells Nanomed Biotechnol. 2016;44(4):1127-32.
30. Hublikar LV, Ganachari SV, Patil VB. Phytofabrication of silver nanoparticles using Averrhoa bilimbi leaf extract for anticancer activity. Nanoscale Adv. 2023;5(16):4149-57. [CrossRef]
31. Giri AK, Jena B, Biswal B, Pradhan AK, Arakha M, Acharya S, et al. Green synthesis and characterization of silver nanoparticles using Eugenia roxburghii DC. Extract and activity against biofilm-producing Bacteria. Sci Rep. 2022;12(1):8383. [CrossRef]
32. Singh P, Mijakovic I. Strong antimicrobial activity of silver nanoparticles obtained by the green synthesis in Viridibacillus sp. Extracts. Front Microbiol. 2022;13:820048. [CrossRef]
33. Ibrahim E, Fouad H, Zhang M, Zhang Y, Qiu W, Yan C, et al. Biosynthesis of silver nanoparticles using endophytic Bacteria and their role in inhibition of rice pathogenic Bacteria and plant growth promotion. RSC Adv. 2019;9(50):29293-9. [CrossRef]
34. Pasieczna-Patkowska S, Cichy M, Flieger J. Application of Fourier transform infrared (FTIR) spectroscopy in characterization of green synthesized nanoparticles. Molecules. 2025;30(3):684. [CrossRef]
35. Sánchez-López E, Gomes D, Esteruelas G, Bonilla L, Lopez-Machado AL, Galindo R, et al. Metal-based nanoparticles as antimicrobial agents:An overview. Nanomaterials. 2020;10(2):292. [CrossRef]
36. Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, Kim JO. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J Biomed Mater Res. 2000;52(4):662-8. [CrossRef]
37. Modi SK, Gaur S, Sengupta M, Singh MS. Mechanistic insights into nanoparticle surface-bacterial membrane interactions in overcoming antibiotic resistance. Front Microbiol. 2023;14:1135579. [CrossRef]
38. Guzman M, Dille J, Godet S. Synthesis and antibacterial activity of silver nanoparticles against gram-positive and gram-negative Bacteria. Nanomedicine. 2012;8(1):37-45. [CrossRef]
39. Wu D, Fan W, Kishen A, Gutmann JL, Fan B. Evaluation of the antibacterial efficacy of silver nanoparticles against Enterococcus faecalis biofilm. J Endod. 2014;40(2):285-90. [CrossRef]