1. INTRODUCTION
Potatoes are of particular importance in feeding humankind [1,2]. This plant is the third staple crop after rice and wheat. Unfortunately, potato is beset by a wide range of different diseases during all developmental stages. Among these diseases, late blight, caused by the oomycete Phytophthora infestans (Mont.) de Bary, is one of the most serious challenges in potato farming. The damage to the global potato industry resulting from late blight amounts to billions of euros per year [3]. vWhile synthetic crop protection chemicals continue to be important in the fight against late blight, many farmers in developing countries cannot afford to use them in potato growing by reason that synthetic agricultural chemicals are somewhat expensive. Additionally, fungicides are toxic to the environment and hazardous to human health, and consumers, especially in developed nations, demand environmentally friendly and safe food products. Therefore, the sustainable intensification of agriculture requires an increased use of genetic solutions instead of synthetic pesticides to protect potatoes against late blight. Such genetic solutions include potato varieties, which have been bred to be resistant to P. infestans infection through introgression of naturally occurring resistance genes (R genes/Rpi genes), which provide the plant with resistance against P. infestans. These genes had been originally discovered in the potato-related wild Solanum species, and some of them were later introduced into potato varieties. R genes belong to the large NB-LRR gene family. Some commercial potato cultivars gained their late blight resistance genes from the wild species S. demissum. In particular, the R1, R2, R3a, R3b, R8, and R9a genes in S. demissum were first characterized and then introduced into potato cultivars [4–9]. To track the introgression of resistance genes into potato cultivars, DNA markers based on polymerase chain reaction (PCR) amplification of the sequences of the corresponding R genes [the so-called sequence-characterized amplified region (SCAR) markers] are traditionally used. These markers are also used in potato breeding for resistance to late blight through marker-assisted selection (MAS).
However, 755 NB-LRR genes were recently found in the genome of the cultivated potato S. tuberosum [10]. Many of these genes are closely related homologues of Rpi genes that arose in ancestral species. In addition, using next-generation sequencing technologies, numerous homologues of the R2 gene were discovered in the Mexican species S. bulbocastanum, S. demissum, S. edinense, S. hjertingii, and S. schenckii [11–13], and homologues of the Rpi-blb1/Rpi-sto1 gene were found in the Mexican species S. bulbocastanum, S. stoloniferum, S. papita, and S. polytrichon [12,14–16]. Moreover, homologues of the Rpi-vnt1 gene originally discovered in S. venturii were found in many South American species of the series Tuberosa [17–20]. At the same time, the functional activity of all these homologues remains unknown, and the identification of functionless homologues of R genes with the SCAR marker technique may cause a lack of correlation between the resistance and the occurrence of these markers. Therefore, to increase the reliability and predictivity of the SCAR markers used, it makes sense to verify the functionality of the target genes based on which these markers are designed. It is advisable to begin checking functionality by assessing the expression (transcription) of these genes because the absence of expression will clearly indicate the absence of function, and this, in turn, will make it possible to immediately reject markers of inactive homologues of R genes.
Thus, the aim of this work was to determine the potential functional activity of late blight resistance genes being revealed with formerly applied SCAR markers in cultivated potato varieties and wild Solanum species. To this end, we estimated the expression of the targets of these markers to subsequently use only those markers in the breeding process, which mark the genes being expressed.
2. MATERIALS AND METHODS
2.1. Overall Experimental Design
The preliminary determination of the functional activity of R genes detected using SCAR markers was carried out by qualitatively determining the presence of transcripts of these genes. To this end, total DNA and total RNA were isolated from the studied plant samples. The total RNA samples were then converted into cDNA in a reverse transcription reaction with random primers. Further, the total DNA samples were screened for 11 SCAR markers, and for those samples in which certain markers were detected, the cDNAs obtained from these samples were analyzed with the same markers. So we could know whether these markers were expressed or not.
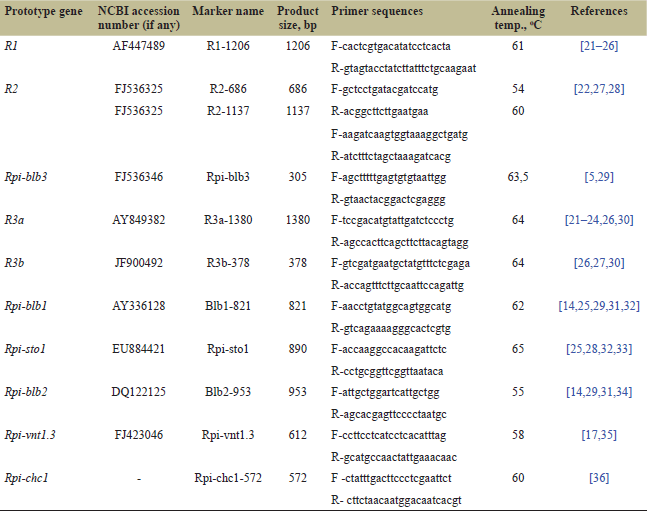 | Table 1. The SCAR markers used in the study. [Click here to view] |
2.2. Plant Material
In this study, we examined plants of nine potato varieties ÒBryanskij krasnyj,Ó ÒGolubizna,Ó ÒZhukovskij rannij,Ó ÒCaprice,Ó ÒLugovskoj,Ó ÒSvitanok kievskij,Ó ÒSudarynya,Ó ÒAlouette,Ó and ÒDesireeÓ as well as plants of two complex interspecific potato hybrids 2359-13 and 2372-60. These varieties and hybrids are supported by the Russian Potato Research Centre. In addition, we used plants of nine accessions of tuber-bearing Solanum species deposited in the N.I. Vavilov Institute of Plant Genetic Resources. These were S. avilesii 20884, S. bulbocastanum 243508, S. bulbocastanum 243512, S. demissum PI161176, S. microdontum k25390, S. okadae 25394, S. polytrichon 24463, S. stoloniferum 24263, and S. stoloniferum 24976. Accordingly, the working collection consisted of 20 samples in total.
2.3. DNA Extraction
Genomic DNA was extracted from the true leaves of young plants with the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the supplier’s instructions. Then, individual DNA samples from eight plants of each accession were blended into one combined sample.
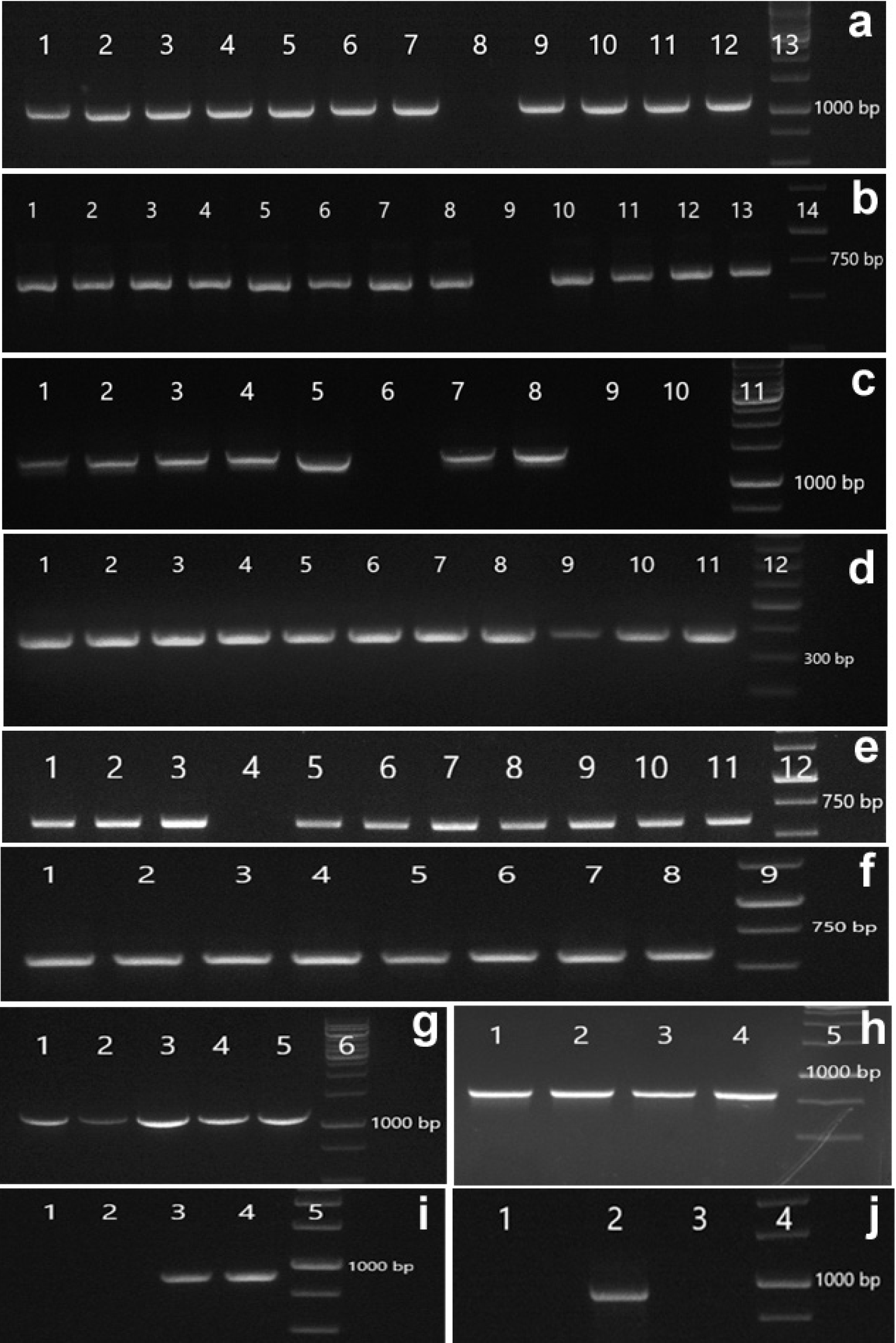 | Figure 1. Results of PCR amplification of cDNA of potato varieties and wild Solanum species with primers of late blight resistance gene markers.* (a) R2-1137: 1- 2359-13, 2- Sudarynya, 3- Svitanok kievskij, 4- 2372-60, 5- Lugovskoj, 6- Brjanskij krasnyj, 7- S. demissum PI 161176, 8- S. stoloniferum 24263, 9- S. bulbocastanum 243512, 10- S. polytrichon 24463, 11- S. bulbocastanum 243508, 12- S. stoloniferum 24976, 13- Gene Ruler 1 Kb DNA Ladder; (b) R2-686: 1- 2359-13, 2- Sudarynya, 3- Svitanok kievskij, 4- 2372-60, 5- Lugovskoj, 6- Brjanskij krasnyj, 7- S. demissum PI 161176, 8- S. stoloniferum 24263, 9- S. microdontum 25390, 10- S. bulbocastanum 243512, 11- S. polytrichon 24463, 12- S. bulbocastanum 243508, 13- S. stoloniferum 24976, 14- Gene Ruler 1 Kb DNA Ladder; (c) R3a-1380: 1- 2359-13, 2- Caprice, 3- Sudarynya, 4- Svitanok kievskij, 5- Alouette, 6- Zhukovskij rannij, 7- 2372-60, 8- Brjanskij krasnyj, 9- S. bulbocastanum 243512, 10- S. polytrichon 24463, 11- Gene Ruler 1 Kb DNA Ladder; (d) R3b-378: 1- 2359-13, 2- Caprice, 3- Sudarynya, 4- Svitanok kievskij, 5- Alouette, 6- Zhukovskij rannij, 7- Golubizna, 8- 2372-60, 9- Brjanskij krasnyj, 10- S. demissum PI 161176, 11- S. stoloniferum 24263, 12- Gene Ruler 100 bp Plus DNA Ladder; (e) Rpi-vnt1.3: 1- Svitanok kievskij, 2- Alouette, 3- Zhukovskij rannij, 4- Lugovskoj, 5- Brjanskij krasnyj, 6- S. demissum PI 161176, 7- S. stoloniferum 24263, 8- S. microdontum 25390, 9- S. bulbocastanum 243512, 10- S. polytrichon 24463, 11- S. okadae 25394, 12- Gene Ruler 1 Kb DNA Ladder; (f) Rpi-chc1-572: 1- 2359-13, 2- Caprice, 3- Sudarynya, 4- 2372-60, 5- Lugovskoj, 6- Brjanskij krasnyj, 7- Desiree, 8- S. avilesii 20884, 9- Gene Ruler 1 Kb DNA Ladder; (g) R1-1206: 1- 2359-13, 2- Svitanok kievskij, 3- Zhukovskij rannij, 4- 2372-60, 5- S. demissum PI 161176, 6- Gene Ruler 1 Kb DNA Ladder; (h) Blb1-821: 1- Sudarynya, 2- S. bulbocastanum 243512, 3- S. bulbocastanum 243508, 4- S. stoloniferum 24976, 5- Gene Ruler 1 Kb DNA Ladder; (i) Rpi-sto1: 1- Sudarynya, 2- S. bulbocastanum 243512, 3- S. bulbocastanum 243508, 4- S. stoloniferum 24976, 5- Gene Ruler 1 Kb DNA Ladder; (j) Blb2 -953: 1- S. stoloniferum 24263, 2- S. bulbocastanum 243512, 3- S. stoloniferum 24976, 4- Gene Ruler 1 Kb DNA Ladder. *Results are presented only for samples that were positive in genomic DNA analysis with the corresponding SCAR markers (see Table 2 [Click here to view] |
2.4. RNA Extraction
Total RNA was extracted from the true leaves of young plants with the ExtractRNA reagent (Evrogen, Russia) according to the supplier’s instructions. Then, individual RNA samples from eight plants of each accession were blended into one combined sample.
2.5. cDNA Synthesis
cDNA preparations were obtained from total RNA samples using Mint reverse transcriptase (Evrogen, Russia) and random primers following the supplier’s instructions.
2.6. SCAR Markers and PCR Conditions
We examined the presence/absence of expression of 10 R genes: R1, R2, Rpi-blb3, R3a, R3b, Rpi-blb1, Rpi-sto1, Rpi-blb2, Rpi-vnt1.3, and Rpi-chc1, detected using 11 SCAR markers of these genes: R1-1206, R2-1137, R2-686, Rpi-blb3, R3a-1380, R3b-378, Blb1-821, Rpi-sto1, Blb2-953, Rpi-vnt1.3, and Rpi-chc1-572 in wild Solanum species and cultivated potato varieties. These markers used to be applied in MAS for breeding resistant to late blight potatoes. Primer sequences for PCR amplification of these markers as well as their annealing temperatures and product size are listed in Table 1. PCR conditions for each of these markers were previously described in the relevant publications (see Table 1), and we used these cycling conditions without changes. The reaction mixture was prepared in a volume of 25 μl; 50 ng of genomic DNA/cDNA was added to each reaction mixture. The thermocycler GeneAmp PCR System 2700 (Applied Biosystems, Inc., USA) was used for PCR amplification. Electrophoresis of PCR products was performed in 1% agarose gel in the presence of ethidium bromide (0.5 μg/ml) in 1X Tris-acetate-EDTA buffer followed by visualization under UV light.
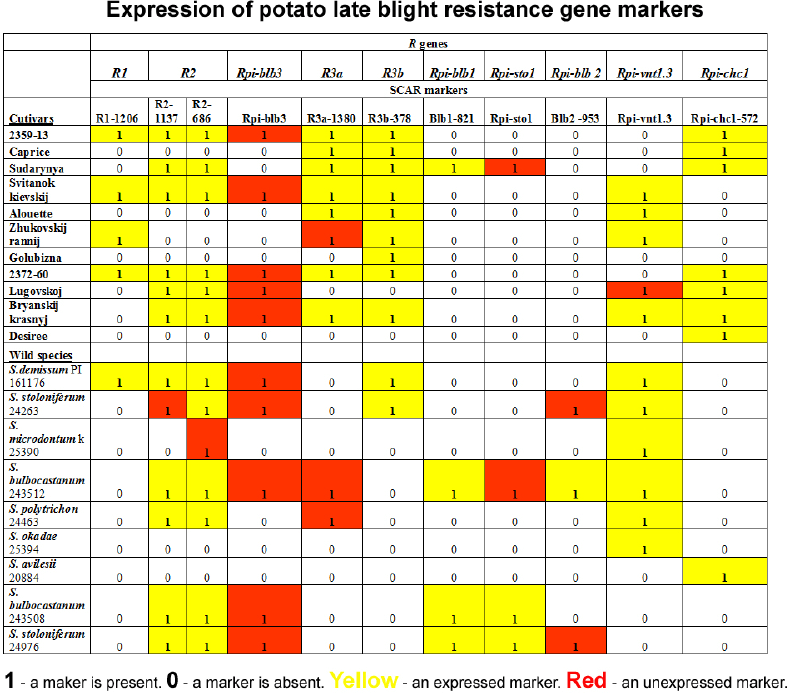 | Table 2. Results of analysis of the expression of potato late blight resistance genes. [Click here to view] |
3. RESULTS
We screened 20 samples of the working collection with 11 SCAR markers of 10 R genes. The results of this screening are presented in Figure 1 and Table 2. In this table, the occurrence or absence of a marker in a sample is indicated as 1 or 0, respectively, while markers of expressed genes are shown on a yellow ground color, and markers of genes for which we were unable to detect expression are shown on a red ground color.
The target genes of markers R1-1206, R2-1137, R2-686, R3b-378, Blb1-821, Rpi-vnt1.3, and Rpi-chc1-572 were expressed in almost all analyzed samples (Table 2). On the other hand, the Rpi-blb3 gene, detected with the Rpi-blb3 marker, was not expressed in any of the samples in which this marker was present.
The R3a gene marker, named R3a-1380, was found in eight potato varieties, and in all of these samples, with the exception of the variety ÒZhukovskij rannij,Ó this gene was expressed. In wild species, this marker was found only in two accessions: S. bulbocastanum 243512 and S. polytrichon 24463, but we failed to find the expression of R3a gene homologues in these samples.
In the matter of the Rpi-blb2 gene, the marker of this gene named Blb2-953 was not detected in potato varieties, but this marker was present in three accessions of wild species: S. bulbocastanum 243512, S. stoloniferum 24263, and S. stoloniferum 24976. However, expression of the Rpi-blb2 gene was observed only in the S. bulbocastanum sample. Both S. stoloniferum samples had no expression of this gene.
We found the Rpi-sto1 gene marker only in the variety ÒSudarynya,Ó while expression of the Rpi-sto1 gene was absent in this cultivar. In wild species, this marker was found in three accessions: S. bulbocastanum 243512, S. bulbocastanum 243508, and S. stoloniferum 24976. The Rpi-sto1 gene predictably was expressed in S. stoloniferum. Speaking of S. bulbocastanum, a homologue of this gene was expressed in S. bulbocastanum 243508 only, and its expression was absent in S. bulbocastanum 243512.
4. DISCUSSION
Marker-assisted selection for resistance to late blight is increasingly being applied in potato farming. Recently, many R gene markers have been developed based on the sequences of these genes. However, because these genes have structural homologues that do not have functional activity, the used markers may detect such homologues, leading to false-positive results when assessing the association of markers with resistance. For example, a homologue of the Rpi-vnt1 gene was found in the late blight-susceptible variety ÒBintje.Ó The authors of the abovementioned research attribute the lack of association between the presence of the Rpi-vnt1 gene and resistance to late blight to the identification of nonfunctional homologues of this gene [37]. In addition, another study [26] showed the absence of a correlation between the levels of resistance to late blight and the number of R gene markers in potato varieties, which, in our opinion, is also due to the fact that the used markers revealed, among other ones, nonfunctional homologues of R genes. In our study, we showed that of the 11 R gene markers analyzed, the markers R1-1206, R2-1137, R2-686, R3b-378, Blb1-821, Rpi-vnt1.3, and Rpi-chc1-572 were expressed in almost all analyzed samples, making these markers good candidates for their further use in MAS for resistance to late blight.
Speaking about the lack of expression of the Rpi-blb3 marker, it should be noted that the Rpi-blb3 marker was originally designed for mapping and cloning the Rpi-blb3 gene [5], and we may conclude that the Rpi-blb3 marker is not suitable for use in breeding process because this marker amplifies an unexpressed target.
Based on the data obtained for the R3a-1380 marker, it can be assumed that outside of S. demissum, the species in which the R3a gene was originally found, homologues of this gene are not expressed and do not have functional activity. Similarly, we can conclude that the Rpi-blb2 gene is not active outside of S. bulbocastanum and the gene from S. bulbocastanum only should be introgressed into potato varieties, but not its homologues from other species. Our data also indicate that the genome of S. bulbocastanum contains both expressed homologues of the Rpi-sto1 gene and unexpressed variants of this gene.
5. CONCLUSIONS
Thus, we analyzed the expression of 10 potato late blight resistance genes that we detected in potato varieties and wild Solanum species using 11 SCAR markers. Our data show that the markers R1-1206, R2-1137, R2-686, R3b-378, Blb1-821, Rpi-vnt1.3, and Rpi-chc1-572 are associated with expressed R genes, and these markers can be recommended for further use in marker-assisted selection for resistance to late blight. Meanwhile, the homologues of the R3a and Rpi-blb2 genes that we identify using the SCAR markers are apparently inactive outside the species in which these genes were originally discovered—S. demissum and S. bulbocastanum, respectively, and the Rpi-sto1 gene has both expressed and non-expressed variants.
6. CONFLICTS OF THE INTEREST
The authors declare that they have no financial or any other competing interests.
7. FUNDING SOURCES
Financial support for this research was provided by the State Task FGUM-2025-0004.
8. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
11. PUBLISHER’S NOTE
All claims expressed in this article are solely those of the authors and do not necessarily represent those of the publisher, the editors and the reviewers. This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
12. USE OF ARTIFICIAL INTELLIGENCE (AI)-ASSISTED TECHNOLOGY
The authors declares that they have not used artificial intelligence (AI)-tools for writing and editing of the manuscript, and no images were manipulated using AI.
REFERENCES
1. Birch PRJ, Bryan GJ, Fenton B, Gilroy EM, Hein I, Jones JT, et al. Crops that feed the world 8: potato: are the trends of increased global production sustainable? Food Secur 2012;4:477–508; CrossRef
2. Bradshaw JE. Review and analysis of limitations in ways to improve conventional potato breeding. Potato Res 2017;60:171–93; CrossRef
3. Haverkort AJ, Boonekamp PM, Hutten R, Jacobsen E, Lotz LAP, Kessel GJT, et al. Durable late blight resistance in potato through dynamic varieties obtained by cisgenesis: scientific and societal advances in the DuRPh project. Potato Res 2016;59:35–66; CrossRef
4. Ballvora A, Ercolano M, Weiss J, Meksem K, Bormann C, Oberhagemann P, et al. The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J 2002;30:361–71; CrossRef
5. Lokossou AA, Park TH, van Arkel G, Arens M, Ruyter-Spira C, Morales J, et al. Exploiting knowledge of R/Avr genes to rapidly clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol Plant Microbe Interact 2009;22:630–41; CrossRef
6. Huang S, van der Vossen EAG, Kuang H, Vleeshouwers VGGA, Zhang N, Borm TJA, et al. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J 2005;42:251–61; CrossRef
7. Li G, Huang S, Guo X, Li Y, Yang Y, Guo Z, et al. Cloning and characterization of R3b; members of the R3 superfamily of late blight resistance genes show sequence and functional divergence. Mol Plant Microbe Interact 2011;24:1132–42; CrossRef
8. Vossen JH, van Arkel G, Bergervoet M, Jo KR, Jacobsen E, Visser RGF. The Solanum demissum R8 late blight resistance gene is an Sw-5 homologue that has been deployed worldwide in late blight resistant varieties. Theor Appl Genet 2016;129:1785–96; CrossRef
9. Jo KR, Visser RGF, Jacobsen E, Vossen JH. Characterisation of the late blight resistance in potato differential MaR9 reveals a qualitative resistance gene, R9a, residing in a cluster of Tm-22 homologs on chromosome IX. Theor Appl Genet 2015;128:931–41; CrossRef
10. Jupe F, Witek K, Verweij W, ?liwka J, Pritchard L, Etherington GJ, et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J 2013;76:530–44; CrossRef
11. Champouret N. Functional genomics of Phytophthora infestans effectors and Solanum resistance genes. Ph.D. thesis, Wageningen University, Wageningen, The Netherlands, 2010.
12. Lokossou AA, Rietman H, Wang M, Krenek P, van der Schoot H, Henken B, et al. Diversity, distribution, and evolution of Solanum bulbocastanum late blight resistance genes. Mol Plant Microbe Interact 2010;23(9):1206–16; CrossRef
13. Aguilera-Galvez C, Champouret N, Rietman H, Lin X, Wouters D, Chu Z, et al. Two different R gene loci co-evolved with Avr2 of Phytophthora infestans and confer distinct resistance specificities in potato. Stud Mycol 2018;89:105–15; CrossRef
14. Wang M, Allefs S, van den Berg RG, Vleeshouwers VG, van der Vossen EA, Vosman B. Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor Appl Genet 2008;116:933–43; CrossRef
15. Lokossou AA. Dissection of the major late blight resistance cluster on potato linkage group IV. Ph.D. thesis, Wageningen University, Wageningen, The Netherlands, 2010.
16. Vleeshouwers V, Raffaele S, Vossen JH, Champouret N, Oliva R, Segretin ME, et al. Understanding and exploiting late blight resistance in the age of effectors. Annu Rev Phytopathol 2011;49:507–31; CrossRef
17. Foster SJ, Park TH, Pel M, Brigneti G, ?liwka J, Jagger L, et al. Rpi-vnt1.1, a Tm-22 homolog from Solanum venturii, confers resistance to potato late blight. Mol Plant Microbe Inter 2009;22:589–600; CrossRef
18. Pel MA, Foster SJ, Park TH, Rietman H, van Arkel G, Jones JDG, et al. Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol Plant Microbe Interact 2009;22:601–15; CrossRef
19. Chen X, Lewandowska D, Armstrong MR, Baker K, Lim JTY, Bayer M, et al. Identification and rapid mapping of a gene conferring broad-spectrum late blight resistance in the diploid potato species Solanum verrucosum through DNA capture technologies. Theor Appl Genet 2018;131:1287–97; CrossRef
20. Paluchowska P, ?liwka J, Yin Z. Late blight resistance genes in potato breeding. Planta 2022;255(6):127; CrossRef
21. Sokolova E, Pankin A, Beketova M, Kuznetsova M, Spiglazova S, Rogozina EV, et al. SCAR markers of the R-genes and germplasm of wild Solanum species for breeding late blight-resistant potato cultivars. Plant Genet Res 2011;9:309–12; CrossRef
22. Sharma R, Kaushik SK, Bhardwaj V, Sharma S, Bhatt AK, Singh BP. Molecular characterisation of potato genotypes for late blight resistance. Potato J 2013;40(2):164–72.
23. Tiwari JK, Siddappa S, Singh BP, Kaushik SK, Chakrabarti SK, Bhardwaj V, et al. Molecular markers for late blight resistance breeding of potato: an update. Plant Breed 2013; 132:237–45; CrossRef
24. Srivastava AK, Singh BP, Kaushik SK, Bhardwaj V, Tiwari JK, Sharma S. Identification of late blight resistance gene homologues in wild Solanum species. Proc Natl Acad Sci India Sect B Biol Sci 2018;88:789–96; CrossRef
25. Gavrilenko TA, Klimenko NS, Antonova OY, Lebedeva VA, Evdokimova ZZ, Gadjiyev NM, et al. Molecular screening of potato varieties bred in the northwestern zone of the Russian Federation. Vavilovskij Zhurnal Genetiki i Selekcii 2018;22(1):35–45; CrossRef
26. Mahfouze HA, El-Sayed OE, Mahfouze SA. Characterization of resistance genes to late blight (Phytophthora infestants) in potato by marker-assisted selection. J Appl Biol Biotech 2023;11(2):178–86; CrossRef
27. Kim HJ, Lee HR, Jo KR, Mortazavian SMM, Huigen DJ, Evenhuis B, et al. Broad spectrum late blight resistance in potato differential set plants MaR8 and MaR9 is conferred by multiple stacked R genes. Theor Appl Genet 2012;124:923–35; CrossRef
28. Lenman M, Ali A, MŸhlenbock P, Carlson-Nilsson U, Liljeroth E, Champouret N, et al. Effector-driven marker development and cloning of resistance genes against Phytophthora infestans in potato breeding clone SW93-1015. Theor Appl Genet 2015;129:105–15; CrossRef
29. Chen S, Borza T, Byun B, Coffin R, Coffin J, Peters R, et al. DNA markers for selection of late blight resistant potato breeding lines. Am J Plant Sci 2017;8(6):1197–209; CrossRef
30. Blatnik E, Horvat M, Berne S, Humar M, Dolni?ar P, Megli? V. Late blight resistance conferred by Rpi-Smira2/R8 in potato genotypes in vitro depends on the genetic background. Plants 2022;11(10):1319; CrossRef
31. Tiwari JK, Devi S, Sharma S, Chandel P, Rawat S, Singh BP. Allele mining in Solanum germplasm: cloning and characterization of RB-homologous gene fragments from late blight resistant wild potato species. Plant Mol Biol Rep 2015;33:1584–98; CrossRef
32. Antonova OY, Klimenko NS, Evdokimova ZZ, Kostina LI, Gavrilenko TA. Finding RB/Rpi-blb1/Rpi-sto1-like sequences in conventionally bred potato varieties. Vavilovskij Zhurnal Genetiki i Selekcii 2018;22(6):693–702; CrossRef
33. Haesaert G, Vossen JH, Custers R, De Loose M, Haverkort A, Heremans B, et al. Transformation of the potato variety Desiree with single or multiple resistance genes increases resistance to late blight under field conditions. Crop Prot 2015;77:163–75; CrossRef
34. Van der Vossen EAGVD, Gros J, Sikkema A, Muskens M, Wouters D, Wolters P, et al. The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J 2005;44:208–22; CrossRef
35. Pel MA. Mapping, isolation and characterization of genes responsible for late blight resistance in potato. Ph.D. thesis. Wageningen University, Wageningen, The Netherlands, 2010.
36. Martynov V, Beketova M. New homologues of the Rpi-chc1 gene in wild and cultivated Solanum species. Crop Breed Appl Biotech 2023;23:e45152337; CrossRef
37. Rogozina EV, Beketova MP, Muratova OA, Kuznetsova MA, Khavkin EE. Stacking resistance genes in multiparental interspecific potato hybrids to anticipate late blight outbreaks. Agronomy 2021;11(1):115; CrossRef