1. INTRODUCTION
Domestic chickens can be found in rural households throughout Southeast and East Asia, making them the most widespread livestock species in the region. Preserving the diversity of chicken genetic resources holds significance in effectively conserving indigenous chicken breeds. In terms of archaeological findings, the earliest undisputed chicken remains can be traced back to the faunal collection at Neolithic Ban Non-Wat, located in central Thailand and are estimated to date from 1650 to 1250 BCE. Following their integration into human communities, chickens spread throughout areas extending beyond the habitats of other Gallus subspecies and species. The advent of rice agriculture potentially played a pivotal role in initiating the process of chicken domestication. Subsequently, the practice of rice agriculture may have facilitated the global dispersal of chickens as they became integrated into human societies [1].
Khiew-Phalee (KP), an indigenous breed of chicken known as Gallus gallus, is a strain of fighting cocks that originated in Uttaradit province, northern Thailand. Its rich history can be traced back to 1770 when it served as the fighting cock of Phraya Phichai Dab Hak, a Siamese general under the rule of King Taksin. Renowned for its exceptional fighting abilities and elegant physique, the KP chicken is hailed as a symbol of Thai warriors. It possesses distinct features such as a glossy blackish-green plumage with visible black shafts around its eyes, beak, neck, back, wings, long curve, and tail. Conservation efforts for this chicken breed have been focused in Phichai district, Uttaradit province, and it received official recognition as a Thai national domestic animal from the Department of Livestock Development in 2013 [2]. Apart from its cultural significance, the KP chicken holds value in esthetic competitions and as a source of recreation and sport.
According to a preliminary study conducted by Phromnoi et al. [3], the KP chicken has been found to possess considerable value. The study revealed that a single KP egg could fetch a price ranging from $5 to $15, whereas a 2-month-old chick could be sold for $45. An 8-month-old mature chick carries a price tag of $90, and an exceptional chick with exceptional genetic attributes could even fetch a remarkable sum of $700 to $900. These transactions predominantly occur in both domestic and international markets, particularly in Middle Eastern countries. The KP chicken represents one of the most valuable assets for the country, as it serves as an agricultural product capable of creating employment opportunities and providing a significant source of income for local communities. Recently, there has been a noticeable decline in the KP chicken population, indicating a concerning trend of decreasing familiarity with this breed.
The physical appearance of the livestock breed can be described by phenotypic traits, as stated by FAO (2003). To ensure the improved conservation and utilization of their genetic resources, the initial step involves conducting a characterization process based on phenotypic and morphological traits, as suggested by AI-Atiyat et al. [4]. Indigenous chicken breeds exhibit significant variations in body conformation, plumage color, and comb type, as well as morphological traits such as feather type, shank color, and ear lobe color. Consequently, the process of characterization provides valuable data regarding the existing and potential future requirements of indigenous populations, while also establishing their current status as distinct breed populations, as highlighted by Tadele et al. [5]. Numerous studies have documented the phenotypic diversity of indigenous chicken breeds in Thailand, including the White Tailed-Yellow Chicken [6], the Black-Bone chicken [7], the Kai Tor-Kai Tang [8], and the KP chicken [9]. By focusing on phenotype-based identification, researchers aim to gain insights into the specific characteristics exhibited by each native chicken breed.
The presence of genetic diversity plays a crucial role in the survival and adaptation of organisms within ever-changing environments. Hence, it is essential to investigate the reproductive connections among animal groups that share common traits or characteristics. As molecular techniques continue to advance, the utilization of nucleotide sequences derived from mitochondrial DNA (mtDNA) has become a valuable biomarker for identifying animal species. This application of mtDNA analysis proves beneficial in various fields such as forensic investigations, studies on animal diversity, and evolutionary research, particularly in constructing phylogenetic trees [10-12]. Nucleotide sequences of a control region (a displacement loop, D-loop), a gene, and a complete genome of mtDNA of several animals are DNA barcoding [13] which has been widely used in species identification, molecular evolution, population genetics, and genetic diversity. The polymorphisms within the sequences of the hypervariable region of the D-loop or control region of mtDNA have made significant contributions to several areas of research and have played a primary role in shedding light on the geographic patterns of genetic diversity and enhancing our understanding of livestock domestication, particularly in the case of chickens. The analysis of polymorphism has provided significant knowledge regarding the evolutionary history and genetic connections among diverse livestock species, contributing to a deeper comprehension of their domestication mechanisms and yielding valuable insights [14,15]. Effective breeding programs are essential for the conservation of the Khiew-Phalee chickens. This study presents for the 1st time the genetic diversity using mtDNA D-loop sequencing information and morphological analysis of KP chickens, providing information about the genetic diversity of these chickens that would be useful for developing and improving breeding strategies. Polymorphism of the D-loop region was evaluated to explore the genetic diversity among KP chickens as well as to highlight the phylogenetic relationship and ancestor of the KP chicken. This research endeavor was conducted to investigate the morphological and genetic variations exhibited by KP chickens by employing a comprehensive approach encompassing morphological characterization techniques and molecular methodologies. The goal is to provide valuable insights as well as data pertaining to breeding enhancements and conservation practices, thereby ensuring the sustained existence and population growth of KP chickens an indigenous avian species that possesses considerable cultural and societal importance within local Thai communities. Moreover, this study has the potential to serve as an exemplary model for safeguarding the nation’s heritage resources, while concurrently exploring avenues for economic viability and profitability through resource development endeavors.
2. MATERIALS AND METHODS
2.1. Research Area
Samples were obtained from a variety of farm locations in Uttaradit province, Thailand, spanning between December 2020 and August 2021. The execution of experimental protocols adhered to the established guidelines set forth by the Animal Ethics Committee at Phibulsongkhram Rajabhat University, situated in Phitsanulok province, Thailand. The study was conducted with full compliance with ethical guidelines, as evidenced by the approval received (Approval reference number: PSRU-(AG)-2020-002).
2.2. Sample Collection
Ethylenediaminetetraacetic acid blood samples were obtained from a selected group of 54 KP fighting cocks, following a purposive sampling approach. These specimens were collected by extracting blood from the wing veins of the birds. The study involved three backyard farms (denoted KB, PP, and KS) located in Uttaradit province, Thailand, which serves as a conservation site for this particular chicken variety. The farm distribution consisted of 14, 9, and 31 birds from the aforementioned farms, respectively.
To obtain total DNA, 20 μL of the collected blood samples were subjected to extraction using the DNeasy® Blood and Tissue Kit (Qiagen, Germany). The DNA extraction process followed the provided manufacturer’s manual. Elution of the DNA was accomplished using 100 μL of sterile nuclease-free water. Subsequently, the eluted DNA samples were carefully stored at a temperature of −20°C until further analysis in the laboratory.
2.3. Selection of Morphological Characteristics for a Phenetic Relationship
Based on the numeric systematics, 14 qualitative morphological characterizations [Table 1] were chosen and employed for the morphological traits of 54 chicken samples. All characteristics were converted into code. For the purpose of cluster analysis, the unweighted pair group method with arithmetic mean technique, as introduced by Patil et al. [16], was utilized. This approach involves grouping entities based on their similarity values. To perform the clustering analysis, the numerical taxonomy system of the multivariate program (NT-SYS) software package version 2.02 [17] was employed. All computations and analysis procedures were executed using this software.
Table 1: Morphological characteristics and code of Khiew-Phalee chicken under observation.
| No. | Character | The state of character and its code |
|---|---|---|
| 1. | Comb | 1=Crested |
| 2. | Ear lobe | 1=Red |
| 3. | Ear hair color | 1=Green-black, 2=Black, 3=Yellow |
| 4. | Nose operculum | 1=Green-black, 2=Black, 3=Yellow |
| 5. | Periocular color | 1=Green-black, 2=Yellow, 3=Marble orange |
| 6. | Beak color | 1=Green-black, 2=Ivory |
| 7. | Hackle color | 1=Green-black, 2=Golden yellow |
| 8. | Back plumage color | 1=Green-black, 2=Gold yellow |
| 9. | Wing plumage color | 1=Green-black, 2=Gold yellow |
| 10. | Underwing plumage color | 1=Green-black, 2=Gold yellow |
| 11. | Flight feather color | 1=Green-black, 2=Black |
| 12. | Main sickle feathers | 1=Green-black, 2=White |
| 13. | Lesser sickle feature | 1=Green-black, 2=Black |
| 14. | Leg/shank/spur color | 1=Green-black, 2=Black, 3=Green |
2.4. Polymerase Chain Reaction (PCR) Amplification and Sequencing
Total DNA extraction from the collected blood involved the utilization of 20 μL and the DNeasy® Blood and Tissue Kit (Qiagen, Germany). All steps and procedures throughout the extraction process strictly adhere to the recommended guidelines provided by the manufacturer. PCR was employed to amplify a specific region of the mtDNA D-loop. This amplification process employed two primers: a forward primer with the sequence CCATCCAACATCTCCGCATGATGAAA [10], and a reverse primer denoted as r2-3m with the sequence 5’ TGCTTAAGGTTAATTACTGCTG 3’ [18]. The amplification process was carried out using the BIORAD T100 Thermocycler (USA). PCR was initiated with an initial denaturation step at 98°C. Subsequently, a total of 35 cycles were performed, each encompassing a denaturation step at 95°C for 30 s, an annealing step at 60°C for 30 s, and an extension step at 72°C for 3 min. Following the cycles, a final extension step was performed at 72°C for 5 min. To verify the length of the amplified fragment, the PCR products were subjected to electrophoresis on a 1.5% agarose gel at 120V for 20 min. Purification of each amplicon was carried out using the FavorPrep GEL/PCR Purification Kit (Favorgen, Taiwan).
Each sample’s amplicon was subjected to amplification using Phusion High-Fidelity DNA polymerase (Thermo Scientific, Lithuania), which possesses proofreading capabilities. The amplicons were then prepared for nucleotide sequence analysis using next-generation sequencing (NGS) based on the MiSeq Illumina sequencing platform. The NGS sequencing process was conducted at BTSeqTM Contiguous Sequencing Service (CELEMICS, South Korea).
2.5. Data Analysis
The ClustalW program, integrated within MEGA version X software, was employed to conduct multiple alignments of nucleotide sequences derived from the D-loop gene of the KP breed, alongside sequences obtained from other breeds sourced from the GenBank database. Haplotype categorization was carried out for specimens obtained from each farm, and various parameters, including haplotype diversity (Hd), DNA Nucleotide diversity models, Pi (the average number of nucleotide differences per site between two sequences), and average number of differences (k), were estimated utilizing DnaSP version 6 software [19]. To determine the nucleotide position of each haplotype, the nucleotide sequence obtained from the Guangxi three-buff chicken (KP681581) was employed. The genetic distance was computed employing the Kimura two-parameter method. Subsequently, the neighbor-joining (Nj) method and unweighted maximum parsimony (MP) tree, with 1000 bootstrap replicates, were employed to construct the phylogenetic tree using MEGA software [20]. A phylogenetic tree analysis was performed following the classification method proposed by Miao et al. [21]. To construct the tree, reference mtDNA sequences from the GenBank database were utilized. These sequences, identified as OK206570-OK206583 (KP-KB 01-14), OK206584-OK206592 (KP-KK 01-09), and OK206594-OK206624 (KP-KS 02-32), had been deposited recently.
3. RESULTS AND DISCUSSION
3.1. Morphological Characteristics
External morphological traits including comb shape, ear lobe color, body plumage color, nose operculum color, periocular color, and color of leg/or shank/spur were assessed for characterization [Figures 1-6]. All KP chickens possessed crested combs, red ear lobes [Figure 1], blackish-green beaks, and blackish-green body plumages [Figure 5]. Variations of morphological traits including ear plumage color, nose operculum color, periocular color, and color of leg or shank were found among subpopulations, dividing the 54 KP chickens into 13 subpopulations or subtypes.
 | Figure 1: Crested comb and red ear lobe. [Click here to view] |
 | Figure 2: Ear plumage (a) blackish green (b) black. [Click here to view] |
 | Figure 3: Periocular color (a) blackish green (b) yellow (c) orange marble. [Click here to view] |
 | Figure 4: Nose operculum and beak color (a) blackish green (b) black (c) yellow. [Click here to view] |
 | Figure 5: Blackish-green color of Khiew-phalee (a) hackle (b) back plumage (c) wing plumage, underwing plumage (e) flight feather, main sickle feathers (g) lesser sickle feature. [Click here to view] |
 | Figure 6: Leg/shank/spur color (a) blackish green (b) black. [Click here to view] |
Among thirteen subpopulations, two different colors of ear plumage were recorded, including blackishgreen (50%) and black (50%) [Figure 2]. Three different colors of the nose operculum were found. The colors were blackish-green (81.48%), black (16.67%), and yellow (1.85%), respectively [Figure 4]. Three different periocular colors were seen, including blackish-green (44.44%), orange marble (29.62%), and yellow (25.93%) [Figure 3]. Like ear plumage, the colors of leg/shank/spur among thirteen subpopulations were blackish-green (88.89%) and black (11.11%) [Figure 6].
In the study, KP chickens were mainly distributed in two subpopulations, including subpopulations 6 (20.37%) (n = 11) and 1 (18.52%) (n = 10), respectively. Both subpopulations had blackish-green plumage throughout the body. The only difference was the orange marble periocular area detected in subpopulation 6. These two populations were distributed across all sampling locations, whereas subpopulations 9–12 were only found in the PP farm, and the proportion was only 1.85–3.7%. Subpopulation 13 only existed on the KS farm (7.4%).
Besides, the NT-SYS-pc tool revealed that two primary clades (clades A and B) were visible among thirteen subpopulations based on morphological data. Meanwhile, the White Tailed-Yellow Chicken (LHK) group was designated as an outgroup. Clade A had four subgroups, whereas Clade B only had two subgroups. The similarity index was found to be between 0.413 and 1.000 (data not shown). Nonetheless, all KP subpopulations (KP-KB, KP-KS, and KP-PP) were reciprocally monophyletic, as shown in Figure 7.
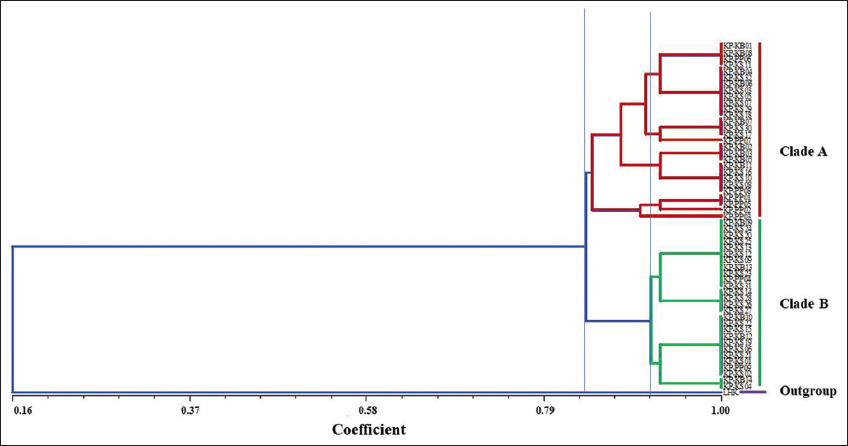 | Figure 7: An unweighted pair group method with arithmetic mean dendrogram of Khiew-Phalee chicken (Gallus gallus) with a simple matching coefficient based on morphological traits. [Click here to view] |
From this study, KP chickens showed high morphological diversity among the population. According to four different morphological traits, including ear plumage color, nose operculum color, periocular color, and leg/shank/spur color. Thirteen subpopulations were described in the study. Blackish-green body plumage (hackle, back, wing, underwing, main sickle feather, flight sickle feather, and lesser sickle feather), a main phenotypic trait of KP chicken, was highly conserved in all subpopulations. AI-Atiyat et al. (2017) postulated that the most discriminate traits are periocular color and shank color. The morphological variation among KP chickens presented in this study might be the result of cross-mating with other subspecies, leading to genetic evolution and changes in morphological characteristics. In addition, the correlation between geographical features and phenotypic traits, such as comb type and earlobe color, has been observed in previous studies [4]. This finding aligns with the current study, where differences in the appearance of certain chickens were noted among the subpopulations originating from three farms located in distinct sites. Information on morphological variation is useful material for the conservation and genetic improvement of KP chickens.
3.2.Molecular Characteristics
The analysis in this study contained 1,232 bp of the mtDNA D-loop gene. The 54 individual KP chickens were submitted to GenBank (Accession numbers OK206570-OK206624). Multiple alignments were used to compare all 54 sequences. Among the 54 samples analyzed, a total of thirty-six nucleotide differences were identified. As indicated in Table 2, the average count of nucleotide differences (k) was determined to be 8.73026. Specifically, one nucleotide deletion was detected at position 859, whereas 35 nucleotide insertions were observed. Most mutations were caused by nucleotide substitution, and 94.28% of these mutations were transitions (34 transition sites and 2 transversion sites). They were categorized into 13 haplotypes (n = 54) with 0.88120 Hd and 0.00679 nucleotide diversity the average number of nucleotide differences per site between two sequences. At KB Farm, a total of three haplotypes (n = 14) were identified, resulting in a Hd of 0.65934, a nucleotide diversity (Pi) of 0.00364, and 16 polymorphic sites (s). The average number of nucleotide differences (k) within this farm was determined to be 5.01099. PP farm displayed a total of four haplotypes (n = 9), resulting in a Hd of 0.75000, a nucleotide diversity (Pi) of 0.00808, and a count of 25 polymorphic sites (s). The average number of nucleotide differences (k) within this farm was calculated to be 10.50000. On the other hand, KS farm exhibited 11 haplotypes (n = 31) with a high Hd of 0.87742, a nucleotide diversity (Pi) of 0.00643, and a total of 29 polymorphic sites (s). The average number of nucleotide differences (k) observed at KS farm was determined to be 8.03441, as presented in Table 2.
Table 2: Genetic diversity of mtDNA D-loop sequence in 54 Khiew-Phalee populations.
| Farm | Number of sequences | Haplotypes | Number of polymorphic sites (s) | Average number of nucleotide differences (k) | Nucleotide diversity (Pi) | Haplotype diversity (Hd) |
|---|---|---|---|---|---|---|
| KB | 14 | 3 | 16 | 5.01099 | 0.00364 | 0.65934 |
| PP | 9 | 4 | 25 | 10.50000 | 0.00808 | 0.62644 |
| KS | 31 | 11 | 29 | 8.03441 | 0.00643 | 0.87742 |
| Total | 54 | 13 | 36 | 8.73026 | 0.00679 | 0.88120 |
As shown in Figure 8, a phylogenetic connection exists among the KP chicken populations using the mitochondrial D-loop region. The complete D-loop nucleotide sequences among the samples in this study and reference samples were analyzed to determine the haplogroups. The nucleotide sequences of the KP population were classified into several haplogroups by the neighbor-joining method with 1,000 bootstrap iterations. Haplotypes 1, 2, 5, 9, and 11 were closely related to haplogroup B. While haplotypes 10 and 12 were related to haplogroup A, haplotypes 8 and 3 were related to haplogroup D. Haplotypes 6 and 4 were related to haplogroups G and F, respectively. Haplotypes 7 and 13 were related to haplogroup E. Haplotype 14 was related to haplogroup C. Correspondingly, the phylogenetic tree in Figure 8, which was constructed using the MP method with 1,000 bootstrap replicates, exhibits the same haplogroup classification as demonstrated.
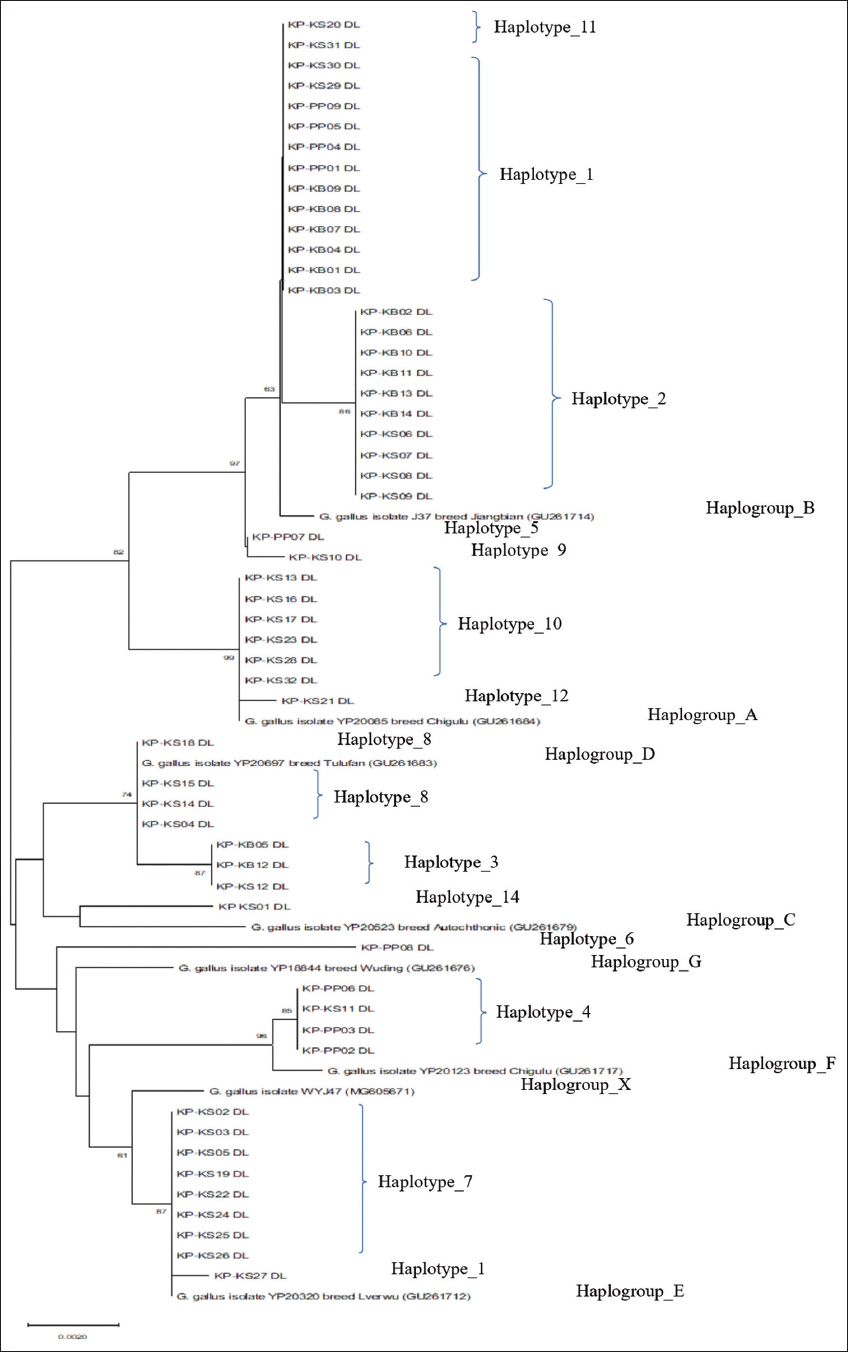 | Figure 8: Phylogenetic tree constructed by D-loop nucleotide sequences of Khiew-Phalee chickens using the neighbor-joining method; 1,000 bootstrap iterations. Black dots represented the haplotypes from this study. [Click here to view] |
The analysis of the mtDNA D-loop of 54 KP chicken samples reveals the presence of 7 haplogroups, suggesting the existence of more than one maternal origin for the chicken populations in the study. Haplogroup B, consisting of three haplotypes including 1, 2, and 11 (n = 24), was the major haplogroup. This haplogroup most likely originated from Southeast Asia (China).
The preservation of genetic diversity is crucial for the long-term viability and adaptability of species within a population. Higher genetic diversity is strongly associated with enhanced population fitness, underscoring the significance of maintaining genetic diversity for conservation purposes. Genetic distance serves as a measure of the genetic variation between or within species in a population. Populations with a multitude of identical alleles exhibit lower genetic distance, indicating a closer genetic relationship and a shared ancestral lineage [22].
For the analysis of genetic diversity in this study, the complete sequences of the mtDNA D-loop region of KP chickens were employed. The D-loop sequences of KP chickens were genetically closely related to other indigenous chickens originating from Asian countries [21]. A high level of genetic diversity was found among KP chickens, as represented by analyzing parameters including s, k, Pi, and Hd [Table 2]. Based on the polymorphism observed in the D-loop region and the number of haplotypes identified, the mtDNA D-loop sequences analyzed in this study displayed notable levels of genetic diversity compared to other chicken breeds in Asia. The findings highlight the substantial genetic resources present within the KP chicken population. This study provides valuable genetic information that can be utilized for the conservation and improvement of breeding programs for KP chickens, contributing to their preservation and enhancement. Phromnoi et al. (2022) recently hypothesized that KP chickens recognized as indigenous Thai fowls, belong to the G. gallus species and exhibit significant similarities to Gallus gallus spadiceus, which is known to originate from China, Myanmar, and Thailand. These findings are congruent with previous research conducted by Teinlek et al. [18], Miao et al. [21], Kawabe et al. [23], Liu et al. [24], Pramual et al. [25], Suwannapoom et al. [26], Xie et al. [27]. These studies also reported that China, India, Laos, Myanmar, and Thailand could be potential origins of indigenous fowls. In terms of genetic diversity and phylogenetic tree, the mtDNA cytochrome B gene indicated five haplotypes from just four polymorphic sites. In this study, the D-loop gene was utilized to classify 13 haplotypes based on 36 polymorphic sites. However, the analysis using D-loop sequences was insufficient to discern differences between haplogroups. Therefore, conducting a comprehensive genome-wide genetic analysis of KP chickens would be beneficial for elucidating their origins and genomic evolution, as well as shedding light on the genetic characteristics of other indigenous chicken breeds. Such investigations would contribute greatly to conservation efforts aimed at preserving these valuable genetic resources.
4. CONCLUSION
According to the study, morphologically complex clusters can be divided into two large clusters, including cluster A (four subpopulations) and cluster B (two subpopulations), although all of the discovered KP subpopulations (KP-KB, KP-KS, and KP-PP) are monophyletic to each other. The majority of individual KP chickens belonged to haplogroup B, according to a study of gene-based genetic variation (mtDNA D-loop). However, KP chicken haplotypes were assigned to haplogroup E and may have recently been influenced by commercial chickens. In addition, substantial morphological variety was found within the KP chicken population, indicating crossbreeding between KP chickens and other subspecies. These findings indicate the importance of breeding the conservation status of this ecological population and considering rebuilding breeding techniques.
This study represents the initial documentation and presentation of genetic diversity through the analysis of polymorphic mtDNA D-loop sequences, alongside the assessment of phenotypic variation in KP chickens. The findings from the current research offer crucial genetic insights that can facilitate the registration of local breeds and enhance the understanding of maternal lineages within the population. Moreover, this information can be utilized to enhance native chicken production performance by implementing crossbreeding strategies involving local and exotic breeds.
5. ACKNOWLEDGMENTS
This work was financially supported by the National Research Council of Thailand Project titled “Biodiversity-Based Economy Development of Khiew-Phalee Native Chicken in Uttaradit for Increasing the Competitive Competence based on Sustainable Utilization and Conservation.” The authors would like to thank Uttaradit Provincial Livestock Office, Thailand, for helping in selecting Khiew-Phalee chickens for the study, as well as the chicken farms that provided blood samples from Khiew-Phalee breeds.
6. AUTHORS’ CONTRIBUTIONS
Interpretation of data; took part in drafting the article All authors made substantial contributions to conception and design, acquisition of data, or analysis and or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
This work was financially supported by the National Research Council of Thailand (grant-NRCT-151/25).
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
Experimental procedures were approved and conducted in accordance with the guidelines of Animal Ethics Committee, Phibulsongkhram Rajabhat University (Approval reference number: PSRU-(AG)-2020-002).
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Peters J, Lebrasseur O, Irving-Pease EK, Paxinos PD, Best J, Smallman R, et al. The biocultural origins and dispersal of domestic chickens. Proc Natl Acad Sci U S A 2022;119:1-9. [CrossRef]
2. Biodiversity Research and Development Section. Khiew-Phalee Native Chicken Web. Available from:https://breeding.dld.go.th/biodiversity/chm/R%20.%20W/-R%20.%20W.html [Last accessed on 2020 Dec 10].
3. Phromnoi S, Ketpiyarat P, Mingchai C, Yeamkong S, Chitmun S, Rodboonsong S, et al. Biodiversity-Based Economy Development of Khiew-Phalee Native Chicken in Uttaradit for Increasing the Competitive Competence Based on Sustainable Utilization and Conservation. [Research Reports]. Thailand:National Research Council of Thailand;2021.
4. AI-Atiyat RM, Aljumaah RS, Abundabos AM, Alotybi MN, Harron RM, Algawaan AS, et al. Differentiation of free-ranging chicken using discriminant analysis of phenotypic traits. Rev Bras Zootec 2017;46:791-9. [CrossRef]
5. Tadele A, Melesse A, Taye M. Phenotypic and morphological characterizations of indigenous chicken populations in Kaffa Zone, South-Western Ethiopia. Anim Husb Dairy Vet Sci 2018;2:1-9. [CrossRef]
6. Yeamkong S, Ngoc Tuan N. Diversity of phenotypic characteristics of White Tailed-Yellow Chicken populations reared under free range system in Phitsanulok Province, Thailand. Biodiversitas 2019;20:1271-80. [CrossRef]
7. Buranawit K, Chailungka C, Wongsunsri C, Laenoi W. Phenotypic characterization of Thai native black-bone chickens indigenous to northern Thailand. Thai J Vet Med 2005;46:547-54. [CrossRef]
8. Incharoen T, Nakhen W, Yeamkong S. Qualitative and quantitative phenotype of Kai Tor-Kai Tang (Gallus gallus) in the lower-northern region of Thailand. Biodiversitas 2022;23:5387-95. [CrossRef]
9. Phromnoi S, Lertwatcharasarakul P, Phattanakunanan S. Genetic diversity and phylogenetic analysis of Khiew-Phalee chickens (Thailand) based on mitochondrial DNA cytochrome b gene sequences. Biodversitas 2022;23:750-6. [CrossRef]
10. Branicki W, Kupiec T, Pawlowski R. Validation of cytochrome b sequence analysis as a method of species identification. J Forensic Sci 2003;48:83-7. [CrossRef]
11. Noro MR, Dubrovo IA, Yoshida MC, Kato M. Molecular phylogenetic inference of the woolly mammoth Mammuthus primigenius, based on complete sequences of mitochondrial cytochrome b and 12S ribosomal RNA genes. J Mol Evol 1998;46:314-26. [CrossRef]
12. Olschewsky A, Hinrichs D. An overview of the use of genotyping techniques for assessing genetic diversity in local farm animal breeds. Animals (Basel) 2021;11:2016. [CrossRef]
13. Peng W, Yang H, Cai K, Zhou L, Tan Z, Wu K. Molecular identification of the Danzhou chicken breed in China using DNA barcoding. Mitochondrial DNA B Resour 2019;4:2459-63. [CrossRef]
14. Adebambo AO, The Chicken Diversity Consortium. Mitochondrial DNA D-loop analysis of Southwestern Nigerian chicken. Arch Zootec 2009;58:637-43. [CrossRef]
15. Likittrakulwong W, Poolprasert P, Roytrakul S. Morphological trait, molecular genetic evidence and proteomic determination of different chickens (Gallus gallus) breeds. J Appl Biol Biotechnol 2019;7:65-70. [CrossRef]
16. Patil P, Sutar S, Malik SK, John J, Yadav S, Bhat KV. Numerical taxonomy of Abelmoschus medik. (Malvaceae) in India. Bangladesh J Plant Taxonomy 2015;22:87-98. [CrossRef]
17. Rohlf FJ. NTSYS-pc:Numerical Taxonomy and Multivariate Analysis System, Version 2.02. Setauket. New York:Exeter Publishing Ltd.;2005.
18. Teinlek P, Siripattarapravat K, Tirawattanawanich C. Genetic diversity analysis of Thai indigenous chickens based on complete sequences of mitochondrial DNA D-loop region. Asian Australas J Anim Sci 2018;31:804-11. [CrossRef]
19. Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6:DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 2017;34:3299-302. [CrossRef]
20. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X:Molecular evolutionary genetics analysis across computing platforms. J Mol Evol 2018;35:1547-9. [CrossRef]
21. Miao YW, Peng MS, Wu GS, Ouyang YN, Yang ZY, Yu N, et al. Chicken domestication:An updated perspective based on mitochondrial genomes. Heredity (Edinb) 2013;110:277-82. [CrossRef]
22. Frankham R. Genetics and extinction. Biol Conserv 2005;126:131-40. [CrossRef]
23. Kawabe K, Worawut R, Taura S, Shimogiri T, Nishida T, Okamoto S. Genetic diversity of mtDNA D-loop polymorphisms in Laotian native fowl populations. Asian Australas J Anim Sci 2014;27:19-23. [CrossRef]
24. Liu YP, Wu GS, Yao YG, Miao YM, Luikart G, Baig M, et al. Multiple maternal origins of chickens:Out of the Asian jungles. Mol Phylogenet Evol 2006;38:12-9. [CrossRef]
25. Pramual P, Meeyen K, Wongpakam K, Klinhom U. Genetic diversity of Thai Native Chicken inferred from mitochondrial DNA sequences. Trop Nat Hist 2013;13:97-106.
26. Suwannapoom C, Wu YJ, Chen X, Adeola AC, Chen J, Wang WZ. Complete mitochondrial genome of the Thai Red Junglefowl (Gallus gallus) and phylogenetic analysis. Zoo Res 2018;39:127-9. [CrossRef]
27. Xie Z, Zhang Y, Deng X, Xie Z, Liu J, Huang L, et al. Molecular characterization of the Cenxi classical three-buff chicken (Gallus gallus domesticus) based on mitochondrial DNA. Mitochondrial DNA A DNA Mapp Seq Anal 2016;27:63968-70. [CrossRef]