1. INTRODUCTION
Phytic acid (PA; C6H18O24P6), formally recognized as myo-inositol-1,2,3,4,5,6-hexakisphosphate (InsP6), boasts a molecular weight of 660 Da. The presence of six phosphorus (P) atoms within the PA molecular structure engenders a negative charge, facilitating its adept binding with positively charged divalent cations, such as iron (Fe), calcium, zinc, and magnesium (Mg). These cations, deemed crucial micronutrients, play pivotal roles in fostering optimal growth and physiological functioning in both humans and animals. By virtue of its chelating prowess as a phytate-metal and phytate-protein complex, PA is renowned for its characterization as an antinutritional compound. PA is profusely prevalent within the soil milieu; however, due to its robust adsorption tendencies onto soil particles, its accessibility for degradation by soil microflora or to plants is substantially restricted [1-3]. A multitude of staple cereal crops, encompassing wheat, rice, maize, sorghum, and pearl millet, are rich reservoirs of vital micronutrients and phosphorus, yet these elements are sequestered by the encumbrance of PA, thereby rendering them unsuitable for direct consumption. In ruminants, the presence of distinct gut microflora endows the capability to degrade and metabolize PA; however, monogastric animals and humans are bereft of this ability [4]. Ameliorating the antinutritional attributes of phytate necessitates the pursuit of alternatives that not only preserve the nutritional integrity of plant-based diets but also amplify the content of phosphorus and micronutrients within edibles and animal feeds. Among the superlative approaches, the application of phytase enzymes stands out; these enzymes are recognized for their ability to catalyze the degradation of PA, liberating inorganic phosphorus and chelated micronutrients for assimilation by both plants and animals.
Augmenting animal feed with phytase not only mitigates the deleterious effects of PA but also contributes to curtailing environmental phosphorus pollution by minimizing the release of unabsorbed phosphorus—a notorious instigator of water body eutrophication. Phytases are widely distributed in nature, with primary sources including plants, microbes (bacteria and fungi), and certain animal tissues [5]. Due to their catalytic properties and ease of enzyme production, microbial-origin phytases are considered the most suitable for commercial biotechnological enzyme production. While some plant roots have been identified with phytases exhibiting low hydrolytic activity and lacking secretion into the rhizosphere, hindering plants’ ability to utilize soil phytates through their mechanisms, microscopic soil fungi actively excrete phytases, with bacteria doing so to a lesser extent. Although plant roots exhibit weak phytase activity, the enzyme’s absence in the rhizosphere prevents plants from independently absorbing fixed phosphorus from soil phytates. Phytases are obtained from microbial sources for commercial applications, owing to the facile manipulation and cultivation of microbial populations. Extracellular phytases have been successfully isolated from diverse fungi, yeasts, and bacteria. Literature abounds with reports detailing the isolation of phytase across various genera, including Bacillus, Pseudomonas, Escherichia, Enterobacter, Klebsiella, Mitsukolla, Selenomonas, Lactobacillus, and others [1,3,6-8].
The current study entails the systematic screening of phytase-producing bacteria within assorted soil samples derived from the northern Gujarat region. This encompasses the morphological and molecular identification of the most promising isolates, followed by the meticulous optimization of physiochemical parameters to obtain the highest enzyme production.
2. MATERIALS AND METHODS
2.1. Chemical Procurement
The acquisition of sodium phytate (sodium salt of PA) was facilitated through HiMedia Chemical Laboratories Pvt. Ltd. Company, Mumbai. All employed chemicals held analytical grade status and were sourced from SRL and Himedia.
2.2. Screening of Phytase-Producing Bacteria
2.2.1. Soil sample collection
A comprehensive collection of soil samples was undertaken across diverse zones within the northern Gujarat region, as suggested by [1]. This encompassed rhizosphere soil from distinct agricultural fields, soil derived from leguminous crop cultivation sites, samples from cattle dung sheds, deposition sites of agricultural and dairy waste, dairy farms, and the micro-forest enclave of the Ganpat University campus.
2.2.2. Media preparation for phytase screening
To discern phytase-producing organisms, a specialized Wheat Bran Extract Agar Media (WBEAM) was deployed, adapting the method proposed by [7]. Notably, wheat bran was employed as an economical substrate replacement for PA. This involved the preparation of wheat bran extract by boiling 20 g of wheat bran in 100 mL of distilled water, followed by filtration through muslin cloth. Furthermore, ammonium sulfate (NH4)2SO4 (0.4 g/L), Mg sulfate heptahydrate (MgSO4.7H2O) (0.2 g/L), casein (1.0 g/L), potassium dihydrogen orthophosphate (KH2PO4) (0.5 g/L), dipotassium hydrogen phosphate (K2HPO4) (0.4 g/L), and agar powder (3%, w/v) were dissolved in 100 mL of distilled water and mixed properly with the wheat bran extract filtrate, eventually making up the volume to 1 L using distilled water. The media was autoclaved at 121°C and 15 psi for 20 min to maintain sterility, and approximately 20 mL of sterilized media was poured into the sterilized petri plates.
2.2.3. Isolation of phytase producers
Soil samples, each weighing 1 g, underwent dilution in 9 mL of 0.1% saline solution, followed by serial dilutions up to 10-6. Subsequently, 100 μL aliquots from each dilution were spread onto WBEAM plates, followed by an incubation period of 48–72 h at 37 ± 1°C. The plates were scrutinized for zones of hydrolysis surrounding the colonies [9].
2.2.4. Confirmation on sodium phytate plates
To confirm the phytase-producing strains, positive results showing hydrolysis of PA colonies were transferred onto Phytase Screening Media plates enriched with sodium phytate (5.0 g/L), ammonium sulfate (NH4)2SO4 (0.4 g/L), Mg sulfate heptahydrate MgSO4·7H2O (0.2 g/L), casein (1.0 g/L), potassium dihydrogen orthophosphate (KH2PO4) (0.5 g/L), dipotassium hydrogen phosphate (K2HPO4) (0.4 g/L), and agar powder (30 g/L) as described by [10]. This step aimed to ascertain the presence of clear halo zones. Further qualitative confirmation of the isolated phytase-producing strains was executed to eliminate false-positive outcomes, following the methodology outlined by Mittal (2011) [11].
2.2.5. Quantitative enzyme assay
The quantification of enzyme production was facilitated through the utilization of WBE broth as the fermentation medium. The microbial culture was inoculated into the fermentation medium, followed by 72 h of incubation under shaking conditions at 150 rpm and 37 ± 1°C. The resultant fermented broth was subjected to centrifugation (at 10,000 rpm for 10 min at room temperature), yielding a supernatant that underwent enzyme precipitation by the addition of equal volumes of absolute ethanol and subsequent overnight incubation at −20°C. The precipitated enzyme, obtained post-centrifugation at 4°C and 9000 g for 15 min, was employed for quantitative assessment using the methodology as established by [12] Sharma and Shukla, 2019.
2.3. Morphological and Biochemical Characteristics
The bacterial isolates were characterized using morphological and biochemical methods, including Gram staining and preliminary morphological identification, following Bergey’s Manual of Systematic Bacteriology. The top two most effective isolates from the four were selected for further molecular analysis.
2.4. Molecular Identification
2.4.1. Isolation of DNA
Two isolates encoded underwent molecular characterization and identification. Cultures were inoculated into nutrient broth and incubated overnight at 37 ± 1°C to facilitate bacterial growth. The incubated broth underwent subsequent centrifugation at 5000 g for 20 min, yielding a pellet that was carefully collected after removing the supernatant. This pellet was then utilized for the genomic DNA extraction using the PowerSoil® DNA Isolation Kit (Qiagen, Germany; Catalogue no.: 12888–50) following the provided protocol.
2.4.2. 16S rRNA sequencing
The quality of the extracted DNA was assessed using a 0.8% low-EEO agarose gel. The genomic DNA was further diluted to a concentration of 100 ng/μL and employed as a template for the amplification of the 16S rRNA gene. The polymerase chain reaction (PCR) was conducted with the forward primer 357F: TACGGGAGGCAGCAG and the reverse primer 1391R: GACGGGCGGTGTGTACA. The following PCR conditions were applied: Initial denaturation at 95°C for 10 min was followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 49°C for 30 s, elongation at 72°C for 90 s, and final extension at 72°C for 7 min. The resulting amplified PCR product was resolved on a 2.0% agarose gel. Subsequently, the DNA sequencing reaction of the PCR amplicon was executed using the same primer set and the BDT v3.1 Cycle Sequencing Kit on the ABI 3500xl Genetic Analyzer. The acquired amplified gene sequences were subjected to BLAST analysis against the available 16S rRNA gene sequence database of the NCBI GenBank. Subsequently, a phylogenetic tree was constructed employing the MEGA software program to facilitate comprehensive analysis and visualization of the evolutionary relationships [13]. The obtained data was also submitted to the NCBI to acquire the accession number.
2.5. Effect of Physio-Chemical Parameters on Phytase Production
In the investigation of phytase production, the two most promising positive isolates, namely CDS and DSS, were employed to elucidate the impact of various physio-chemical parameters on enzyme production. The study encompassed a meticulous assessment of factors such as inoculum percentage, fermentation time, the initial pH of the fermentation medium, and the optimal fermentation temperature. For each parameter examined, enzymatic assays were meticulously conducted following the methodology outlined above, ensuring rigorous scientific precision. Furthermore, to ascertain the robustness of the findings, all experiments were conscientiously repeated in triplicate (n = 3), thereby enhancing the statistical reliability of the observations.
2.5.1. Effect of inoculum percentage on phytase production
In the exploration of the impact of inoculum concentration on phytase production, the fermentation media flasks were subjected to inoculation with diverse concentrations of the 24-hour-old inoculum. These concentrations encompassed percentages of 1%, 5%, 10%, 15%, 20%, and 25%. The fermentation broths were incubated at 37°C and 150 rpm for 72 h. Upon the culmination of the incubation, the fermented media were judiciously sampled to conduct rigorous enzymatic assays.
2.5.2. Effect of time on phytase production
In pursuit of understanding the temporal dynamics of phytase production, the fermentation media underwent inoculation with a 24-hour-old broth culture. These cultures were then incubated at 37°C under continuous shaking at 150 rpm to stimulate cellular phytase synthesis. At defined intervals of 24 h, ranging up to 5 days, 20 mL of the fermentation broth were judiciously harvested. The harvested media was subjected to centrifugation, followed by careful collection of the supernatant, which was subsequently subjected to meticulous estimation via the phytase assay.
2.5.3. Effect of temperature and pH on phytase production
The impact of temperature and pH on phytase production was probed by preparing distinct fermentation broths in separate flasks, each corresponding to a specific pH value (ranging from pH 4 to pH 9) and temperature (spanning room temperature, 37°C, 40°C, 45°C, 50°C, and 55°C). To achieve the desired pH levels, precise adjustments were made using 0.1 N HCl and 0.1 N NaOH. Subsequent to a 3-day incubation period under constant agitation at 150 rpm, the fermented broth was meticulously collected, followed by centrifugation. The supernatant was then systematically gathered, and phytase activity was quantitatively assessed employing the established routine methodology. Through this systematic exploration of the diverse physio-chemical parameters, the study seeks to contribute valuable insights into the modulation of phytase production, facilitating a more comprehensive understanding of its underlying mechanisms and potential applications.
3. RESULTS
In earlier studies, phytase enzyme-producing bacterial isolates were screened and identified from various sources, including poultry farms, dairy areas, hydrothermal vents, hot springs, agricultural soil, etc. [7,13-19]. In the present study, screening of phytase-producing bacterial isolates obtained from soil samples in the north Gujarat region was conducted. A total of 42 colonies were identified, displaying clear zones of hydrolysis around them, indicating phytase production [Figure 1].
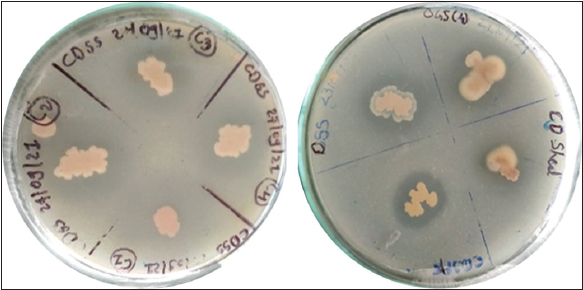 | Figure 1: Screening of bacterial isolates for enzyme production based on zone of hydrolysis. [Click here to view] |
Subsequently, the top 13 isolates demonstrating hydrolysis efficiency exceeding 50% were selected for further investigation via quantitative enzymatic assays [Figure 2]. Based on both plate assays and quantitative enzymatic assays, two isolates, DSS and CDS, exhibited significantly higher phytase activity compared to the other isolates [Table 1]. Morphological analysis confirmed that both DSS and CDS isolates were gram-positive [Table 2].
 | Figure 2: Phytase activity of the various isolates screened. [Click here to view] |
Table 1: Hydrolysis efficiency of isolates.
| S. No. | Isolates | Zone diameter (mm) | Colony diameter (mm) | Hydrolysis efficiency (%), ((Z-C/C) *100) |
|---|---|---|---|---|
| 1 | ASC | 20 | 8 | 150.00 |
| 2 | BGS-1 | 10 | 5 | 100.00 |
| 3 | CDS | 30 | 11 | 172.73 |
| 4 | CDSAS | 15 | 7 | 114.29 |
| 5 | CDSS | 28 | 11 | 154.54 |
| 6 | CDSS-2 | 15 | 7 | 114.29 |
| 7 | DSS | 42 | 14 | 200 |
| 8 | OGS-1 | 10 | 5 | 100 |
| 9 | OGS2.2 | 32 | 14 | 128.57 |
| 10 | BGS4 | 10 | 4 | 150 |
| 11 | MP1 | 10 | 5 | 100 |
| 12 | MF4 | 10 | 4 | 150 |
| 13 | LGS-1 | 13 | 6 | 116 |
CDSS: Clinical decision support system
Table 2: Morphological properties of the best isolates.
| S. No. | Isolates/tests | CDS | DSS |
|---|---|---|---|
| 1 | Gram’s staining | + | + |
| 2 | Size | Big | Small |
| 3 | Shape | Irregular | Round |
| 4 | Margin | Wavy/Undulated | Entire |
| 5 | Elevation | Flat | Flat |
| 6 | Surface | Rough | Smooth |
| 7 | Consistency | Wet and soaked | Moist |
| 8 | Odour | Yes | No |
| 9 | Optical characteristics | Translucent | Translucent and white |
| 10 | Pigmentation | Pink | No |
DSS: Decision support system, CDS: Clinical decision support
To identify these isolates, 16S rRNA sequencing was performed [Figure 3]. Based on the bioinformatic analysis, the isolates DSS and CDS were found to be Bacillus tequliensis [Figure 4] and Bacillus spp. [Figure 5]. Sequences were submitted to obtain an accession number, and the NCBI database provided unique accession numbers OP412485 and OR142655 for DSS and CDS, respectively.
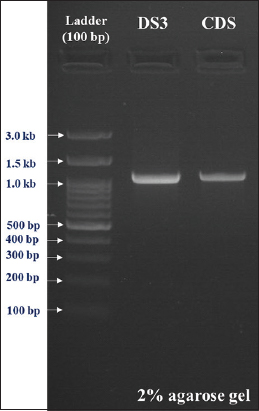 | Figure 3: Amplification of 16S rRNA gene. [Click here to view] |
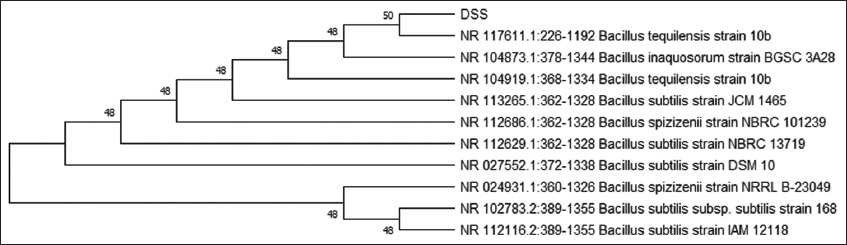 | Figure 4: Phylogenetic tree analysis of decision support system. [Click here to view] |
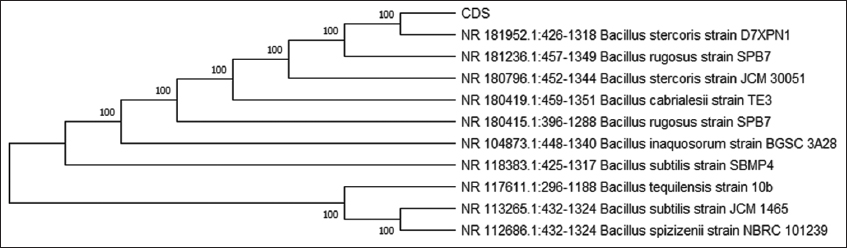 | Figure 5: Phylogenetic tree analysis of clinical decision support. [Click here to view] |
The primary objective of the optimization efforts was to determine the optimal physiochemical conditions for achieving the highest phytase yield. Earlier studies have reported variations in maximum phytase production both among genera and within species of the same genus. For instance, incubation times vary, with Enterobacter, Pseudomonas fluorescens, and Bacillus typically requiring 72 h, while Aspergillus heteromorphus thrives best at 120 h of incubation. Additionally, different strains of the Bacillus genus have demonstrated varying optimal enzyme production times, ranging from 48 to 96 h [7,20-26].
In our study, isolates DSS and CDS also exhibited peak enzyme production after 72 h of fermentation, after which enzyme activity began to decline [Figure 6a]. This finding aligns with previous observations and underscores the importance of optimizing incubation times for specific strains to maximize phytase production.
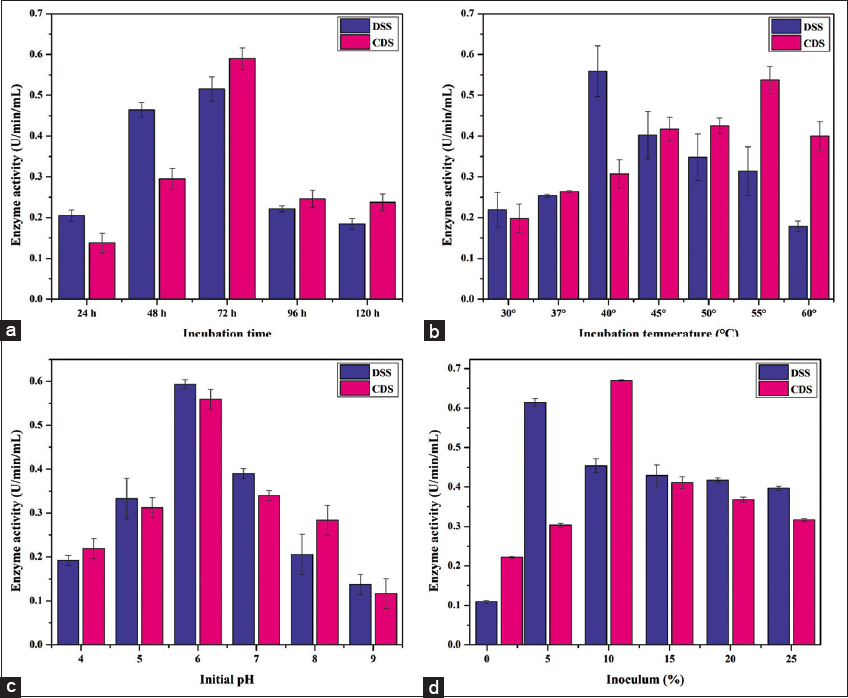 | Figure 6: Effect of Incubation time (a), Incubation temperature (b), initial pH (c) and inoculum perecntage (d) on phytase production. [Click here to view] |
Reports concerning the optimal temperature for the growth and enzyme production of bacterial isolates indicate a wide temperature range, spanning from 28°C to 65°C. Previous studies have revealed specific temperature preferences for different bacterial strains. For example, thermotolerant Klebsiella has been observed to achieve maximum enzyme production at 55°C, while Bacillus strains S1 and S2 exhibit peak enzyme production at 40°C [17]. Bacillus isolate C43 achieves its maximum enzyme production at 45°C [7], Bacillus tequiliensis at 60°C [17], and Bacillus HCYL03 at 45°C [21]. In this study, despite both isolates being Bacillus strains, we observed a temperature preference difference. Specifically, isolate DSS displayed maximum enzyme production at 40°C, while isolate CDS exhibited its peak enzyme production at 55°C [Figure 6b]. This temperature divergence emphasizes the importance of tailoring the fermentation conditions to the specific characteristics of each isolate to optimize enzyme production.
The initial pH of the fermentation medium is another critical factor influencing metabolite production. Earlier reports have indicated that phytase-producing bacteria can thrive in pH ranges spanning from 5 to 7. However, maximum enzyme production is typically achieved at specific pH levels. For instance, Klebsiella spp. has been found to produce phytase in an acidic medium, while Bacillus strains US417, S1, and S2 have reported pH 6.5 as optimal for maximum enzyme production. In contrast, Bacillus subtilis MJA from soil has shown an optimum pH of 7 for enzyme production [17,25,27]. In this experimental work, both the isolates exhibited the highest enzyme activity at pH 6 [Figure 6c], aligning with the trend observed for some Bacillus strains. This finding highlights the significance of adjusting the pH of the fermentation medium to promote optimal enzyme production in specific isolates.
Studies have demonstrated that the production of phytase increases as the percentage of inoculum increases. The maximum concentration of inoculum that affects enzyme production has been reported to be 10% in some cases [7]. However, for fungal cultures, 2% has been identified as the optimized inoculum concentration [23]. In our research, we found that a 5% inoculum played a crucial role in maximizing enzyme production for isolate DSS, whereas for CDS, the highest enzyme activity was achieved with a 10% inoculum [Figure 6d]. These findings underscore the importance of considering inoculum concentration as a key factor in optimizing enzyme production for different bacterial isolates.
4. DISCUSSION
To achieve higher enzyme yields, it is crucial to provide microorganisms with optimal cultural conditions, encompassing physical, environmental, and nutritional requirements. Any alterations in the growth conditions, such as changes in temperature, pH, fermentation duration, inoculum concentration, or nutritional needs, can significantly impact the metabolic activity of the microorganism and, consequently, enzyme production. It’s important to note that the effects of these variations can vary widely between different species and even among isolates of the same species. Therefore, optimizing fermentation parameters is essential to develop economically viable technologies for maximum phytase production.
Incubation time is a critical factor influencing maximum enzyme production. Previous studies have demonstrated that maximum enzyme production was achieved when maintaining a temperature of 55°C for CDS and 40°C for DSS. Similarly, the highest phytase production was reported at pH 6 [Figure 6c]. It’s worth noting that enzyme production declined progressively above 50°C, and enzyme production decreased at pH levels above 8. Several reports have indicated that 72 h is the optimal incubation time for maximum enzyme production, a finding supported by our results, which also showed that phytase activity peaked at 72 h of fermentation [Figure 6a]. Extending the fermentation duration beyond this point led to reduced enzyme production and phytase activity, possibly due to proteolytic enzyme degradation or the accumulation of secondary metabolites.
Each microbial isolate possesses a unique combination of conditions that promote higher enzyme production. As studies have suggested, Bacillus C43 exhibits higher production at pH 6, 45°C for 72 h with a 10% inoculum [8], while Bacillus strain HCYL03 achieves optimal conditions at pH 5, 45°C, and 72 h [21].
In our study, we determined that pH 6 and 72 h were suitable for maximum enzyme production for both isolates. However, the temperature and inoculum percentage varied between the isolates. Specifically, DSS demonstrated higher enzyme activity when grown at temperatures ranging from 37°C to 40°C with a 5% inoculum, while CDS displayed peak activity at 45°C with a 10% inoculum, effectively stimulating phytase production. In conclusion, we have identified two new phytase-producing strains and determined the best physiochemical condition for optimal phytase production. Further study is undergoing to find out the optimum nutrient requirement for maximum enzyme production based on the one variable at a time approach.
5. CONCLUSION
In the current study, we identified the two potential isolates for the phytase production, in which enhancement of enzyme production was observed when physiological parameters were optimized. Based on the phylogenetic analysis both the isolates were characterised under the genus Bacillus.
6. AUTHORS’ CONTRIBUTIONS
Madhvi contributed to design, acquisition, analysis and interpretation of data; drafting the article. Madhvi and Hardik Shah revising it critically for important intellectual content and approved the final version; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be authors as per the International Committee of Medical Journal Editors (ICMJE) requirements and guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Prajapati M, Shah H. Impacts and industrial applications of phytic acid and phytase J Pure Appl Microbiol 2022;16:2292-302. [CrossRef]
2. Perera I, Seneweera S, Hirotsu N. Manipulating the phytic acid content of rice grain toward improving micronutrient bioavailability. Rice (NY) 2018;11:4. [CrossRef]
3. Liu X, Han R, Cao Y, Turner BL, Ma LQ. Enhancing phytate availability in soils and phytate-P acquisition by plants:A review. Environ Sci Technol 2022;56:9196-219. [CrossRef]
4. Naghshbandi MP, Moghimi H. Stabilization of phytase on multi-walled carbon nanotubes via covalent immobilization Methods Enzymol 2020;630:431. [CrossRef]
5. Konietzny U, Greiner R. Molecular and catalytic properties of phytate-degrading enzymes (phytases). Int J Food Sci Technol 2002;37:791-812. [CrossRef]
6. Thakor R, Mistry H, Bariya H. Isolation and identification of nitrogen fixing bacteria:Azoarcus species. In:Amaresan N, Patel P, Amin D, editors. Practical Handbook on Agricultural Microbiology. Humana, New York:Springer Protoc. Handbooks;2022. [CrossRef]
7. Rizwanuddin S, Kumar V, Singh P, Naik B, Mishra S, Chauhan M, et al. Insight into phytase-producing microorganisms for phytate solubilization and soil sustainability. Front Microbiol 2023;14:1127249. [CrossRef]
8. Sreedevi S, Reddy BN. Screening for efficient phytase producing bacterial strains from different soils. Int J Biosci 2013;3:76-85.
9. Han X, Ding S, Lu J, Li Y. Global, regional, and national burdens of common micronutrient deficiencies from 1990 to 2019:A secondary trend analysis based on the global burden of disease 2019 study. EClinicalMedicine 2022;44:101299. [CrossRef]
10. Singh P, Kumar V, Agrawal S. Evaluation of phytase producing bacteria for their plant growth promoting activities. Int J Microbiol 2014;2014:426483. [CrossRef]
11. Mittal A. Isolation and biochemical characterization of acido-thermophilic extracellular phytase producing bacterial strain for potential application in poultry feed. Jundishapur J Microbiol 2011;4:273-82.
12. Sharma B, Shukla G. Isolation, identification, and characterization of phytase producing probiotic lactic acid bacteria from neonatal fecal samples having dephytinization activity isolation, Identification, and characterization of phytase producing probiotic lactic acid. Food Biotechnol 2020;34:151-71. [CrossRef]
13. Singh, M. Kumar, A. Mittal, Mehta PK. Microbial enzymes:Industrial progress in 21st century. 3 Biotech 2016;6:1-15. [CrossRef]
14. Widawati S, Ikhwani AZ, Sudiana IM. Phytase activity of phytase-producing bacteria isolated from mangrove sediment. IOP Conf Ser Earth Environ Sci 2022;976:012041. [CrossRef]
15. Lin H, Han T, Wang J, Ma Z, Yu X. Screening and identification of a strain with protease and phytase activities and its application in soybean meal fermentation. Appl Biochem Biotechnol 2023;19:1-4.
16. Nezhad NG, Rahman RN, Normi YM, Oslan SN, Shariff FM, Leow TC. Isolation, screening and molecular characterization of phytase-producing microorganisms to discover the novel phytase. Biologia (Bratisl) 2023;2527-37. [CrossRef]
17. Badoei-Dalfard A, Parhamfar M, Karami Z. Characterization of a thermostable, acidic-phytase from Bacillus tequilensis Dm018;medium optimization by response surface methodology. Catal Lett 2019;149:2961-72. [CrossRef]
18. Muslim SN, Mohammed Ali AN, Al-Kadmy IMS, Khazaal SS, Ibrahim SA, Al-Saryi NA, et al. Screening, nutritional optimization and purification for phytase produced by Enterobacter aerogenes and its role in enhancement of hydrocarbons degradation and biofilm inhibition. Microb Pathog 2018;115:159-67. [CrossRef]
19. Alias N, Shunmugam S, Ong PY. Isolation and molecular characterization of phytase producing bacteria from Malaysia hot spring. J Fundam Appl Sci 2018;9:852. [CrossRef]
20. Tungala A, Narayanan KA, Muthuraman MS. Isolation of phytase producing bacteria from poultry faeces and optimization of culture conditions for enhanced phytase production. Int J Pharmacy Pharm Sci 2013;5:3-8.
21. Sardar R. Optimization of phytase production by Bacillus sp. (HCYL03) under solid-state fermentation by using box-behnken design. Braz Arch Boil Technol 2022;65. [CrossRef]
22. Rani R, Ghosh S. Production of phytase under solid-state fermentation using Rhizopus oryzae:Novel strain improvement approach and studies on purification and characterization. Bioresour Technol 2011;102:10641-9. [CrossRef]
23. Lata S, Rastogi S, Kapoor A, Imran M. Optimization of culture conditions for the production of phytase from Aspergillus heteromorphus MTCC 10685. Int J Adv Biotechnol Res 2013;4:224-35.
24. More SB, Khan S, More SM, Girde AV, Shinde VA. Phytase production by Bacillus species and optimization of fermentation parameters. Indian J. Microbiol 2014;17:7-14.
25. Kammoun R, Farhat A, Chouayekh H, Bouchaala K. Phytase production by Bacillus subtilis US417 in submerged and solid state fermentations. Ann Microbiol 2012;62:155-64. [CrossRef]
26. Liu L, Li A, Chen J, Su Y, Li Y, Ma S. Isolation of a phytase-producing bacterial strain from agricultural soil and its characterization and application as an effective eco-friendly phosphate solubilizing bioinoculant isolation of a phytase-pro. Commun Soil Sci Plant Anal 2018;3624. [CrossRef]
27. El-toukhy NM, Youssef AS, Mikhail MG. Isolation, purification and characterization of phytase from Bacillus subtilis MJA. Afr J Biotechnol 2013;12:2957-67.