1. INTRODUCTION
Lactoferrin is an 80-kDa protein with 703 amino acids that belongs to the transferrin family [1,2]. It is made up of a single polypeptide chain that has been folded into two lobes (the N-lobe and the C-lobe) [2-5]. Lactoferrin can be found in milk, tears, nasal secretions, saliva, urine, uterine secretions, and amniotic fluids, as well as in secondary granules of neutrophils [2,6-8]. Lactoferrin’s unique ability to bind to various cell surface receptors in different tissues is one of its distinguishing characteristics. Lactoferrin’s ability to bind is one of the reasons it has so many health benefits, including immune system regulation, tumor and cancer prevention, safely treating anemia and osteoporosis by adjusting homeostasis, and acting as a natural antibacterial and antioxidant supplement.
High-cell density fermentation (HCDF) is a critical strategy in the large-scale engineering of host strains for industrial protein, peptide, and amino acid production [9]. Using the HCDF technique increases protein productivity, improves downstream yield, decreases culture volume, and reduces production costs. Fed-batch fermentation is a cost-effective operational strategy for HCDF [10]. Pichia pastoris fed-batch cultivation is divided into three phases: The glycerol batch phase (S1), the nutrient fed-batch phase (S2), and the methanol fed-batch phase (S3) [11,12]. The S1 and S2 phases are known as the biomass production phases, and their goal is to maximize cell density. The S3 phase is known as the induction phase because it is designed to stimulate the synthesis of the target protein or product. Feeding strategies include constant feeding rate, increased feeding rate, exponential feeding rate, pH-stat, DO-stat, and μ-stat, among others [12]. Good cell density and recombinant protein productivity could be obtained by using a suitable fed-batch fermentation strategy that separated the cell growth phase, protein production phase, and induction time. Using optimal feeding strategies, the dry cell weight of P. pastoris could reach 65.8 g/L for production of Saccharomyces cerevisiae L-Asparaginase II [13] or OD600nm of P. pastoris reached around 400 for production of a fungus β-glactosidase [12]. Additional attributes, including the yields of heterologous proteins, could reach 22 g/L for the production of the recombinant hydroxynitrile lyase [14], or 14.8 g/L for the production of the recombinant gelatins [15].
P. pastoris has received a lot of attention for being a powerful system for expressing hundreds of proteins from viruses, bacteria, fungi, animals, plants, and humans [12,16]. P. pastoris is a methylotrophic yeast with eukaryotic machinery that grows on methanol as its sole carbon source. The genes are integrated into the genome via the alcohol oxidase I promoter, resulting in exogenous overexpression of recombinant proteins in P. pastoris. Lf from humans, horses, pigs, goats, cows, and sheep has been expressed in a variety of hosts, including yeast and cows [17-19]. The obtained expression levels range between 2 and 1200 mg/L, and the biological activities have been evaluated [18]. However, this recombinant protein has only been produced in flasks; production on a larger scale, such as in a 2-L or 10-L bioreactor, has not been reported.
Previously, we successfully generated and expressed a recombinant construct, pPICZA: bLfopt, carrying the gene encoding bovine lactoferrin (bLf) in P. pastoris [20]. The medium and conditions were also chosen and optimized for lactoferrin production from the P. pastoris KM71H-3 strain in a 250-mL flask [21]. However, the cell density was still low at this scale, with an OD600 of around 50, corresponding to the low lactoferrin content. There has been little research into the large-scale production of recombinant lactoferrin in P. pastoris. More research is needed to provide enough data to enrich research on the large-scale production of P. pastoris lactoferrin. This study focused on developing the fermentation strategy to obtain the high-cell density of the P. pastoris KM71H3 strain for the production of bLf by using several nutrient-feeding strategies. The P. pastoris strain was first fermented in a 2-L bioreactor with the same medium and optimized conditions on the flask scale. The feeding strategies with glycerol solution and glycerol-mixed nitrogen sources were then investigated in the S2 phase. The fermentation process was then scaled up to the 10-L and 100-L bioreactors. Finally, methanol feeding strategies were investigated to achieve high lactoferrin levels. As a result, the study provided a starting point for future industrial production of recombinant lactoferrin in P. pastoris.
2. MATERIALS AND METHODS
2.1. Strain and Media
The P. pastoris KM71H-3 strain carrying the gene coding for bLf (pPICZαA: :bLfopt) was constructed in our laboratory by transforming the recombinant plasmid pPICZαA: :bLfopt into the host P. pastoris KM71H strain.
Fermentation medium (w/v): 17 g/L KH2PO4, 15 g/L (NH4)2SO4, 2 g/L MgSO4·7H2O; 100 mL/L phosphate buffer 1M, 5 g/L corn steep, and 10 mL/L glycerol 100%.
Trace salts PTM4 (w/v): CuSO4.5H2O 0.2%; NaI 0.008%; MnSO4. H2O 0.3%; (NH4)6Mo7O24.4H2O 0.0148%; H3BO3 0.002%; CoCl2 0.05%; ZnCl2 0.7%; FeSO4.7H2O 2.2%; Biotin 0.02%; H2SO4 0.1%. PTM 500X was prepared as a stock solution.
Feeding solution:
Glycerol feeding solution: 30% glycerol (w/v); PTM4 ×500 salts and Biotin ×500 were added to ×1 in the final feeding solution
Glycerol, nitrogen - source feeding solution: 30% glycerol (w/v), 2.62% corn-steep, 9.2% (NH4)2SO4 and PTM4 ×500 salts, Biotin ×500
Methanol solution: 50% methanol (w/v) and PTM4 ×500 salts, Biotin ×500.
2.2. Seed Cultivation
A single colony from the YPD plate was used to cultivate in 5 mL of BMGY-bleomycine at 28°C and 250 rpm for 18 h. The preculture was then transferred to the 100 mL or 500 mL of BMGY medium, depending on the scale of batch fermentation in the bioreactors and then was transferred to the bioreactor. The inoculum volume was then adjusted to ensure an initial biomass concentration corresponding to an OD600 of 0.5.
2.3. Batch Fermentation
Batch cultivations were carried out in a 2-L bioreactor (Sartorius 1601, Germany) with a 1-L working volume. The basal salt medium was sterilized in the bioreactor, and pH was adjusted to 6.0 by H3PO4 and NH4OH. Sterile-filtered trace elements were transferred to the bioreactor. The inoculum volume was adjusted to ensure an initial biomass concentration corresponding to an OD600 of 0.5. The bioreactor was programmed to maintain the pH at 6.0, the temperature at 28°C, and the pO2 at 20%. Temperature, pH, and pO2 as well as agitation were recorded online. The fermenter was supplied with a 50% MeOH solution to ensure that the MeOH concentration was 0.5% every 24 h. The pumping rate when inducing MeOH was set to ensure the concentration of methanol was at 0.5% for 24 h. 10 mL of broth samples were taken regularly every 12 h to measure biomass (OD600) and protein concentration.
2.4. Fed-Batch Fermentation
The medium and inoculum were prepared as for the batch fermentation. The inoculum volume was adjusted to ensure an initial biomass concentration corresponding to an OD600 of 0.5. The batch phase was performed in 15 h, and then the glycerol-fed-batch fermentation was performed as previously reported [17,22].
In the glycerol fed-batch stage, a feeding solution was added after 15 h of fermentation, ensuring a final glycerol concentration of 1% in the medium. All fermentation conditions, including temperature, pH, DO, and anti-foam agent, were the same as those in the previous phase.
In the methanol induction phase, methanol solution was supplemented into fermenters and inducted for 50 h. The methanol solution was added once or continuously to ensure a concentration of 0.5% for 24 h. Broth samples were taken hourly during the fermentation process to measure the biomass and protein concentration.
2.5. Sample Analysis
The samples were continuously taken every 2 h from the fermentation broth. The growth of P. pastoris during the fermentation was determined by measuring the absorbance of the broth culture at 600 nm. Serial 5-mL samples of broth cultures diluted to OD600 values ranging from 10 to 60 were prepared. Wet cell weight and dry cell weight were determined by centrifugation of 5-mL samples (5000 rpm, 4°C, and 10 min) in a centrifuge; the pellet was washed twice with 5 mL of deionized water; the wet cell weight was estimated; following that, subsequent oven drying at 95°C to a constant weight was performed to calculate the dry cell weight. Finally, a calibration curve was generated to show the relationship between the absorbance and dry cell weight as well as wet cell weight.
2.6. Protein Extraction by Sonicator
After induction, the cells were harvested by centrifugation before being dissolved in phosphate buffered saline (PBS) buffer. The solution was then sonicated using a 2-mm probe sonicator for 600 s (10 s on and 10 s off) at 60% power. Following that, the samples were put on ice. Then, the samples were centrifuged to discard the broken cells, while the supernatant was retained for further analysis.
2.7. Dot Blotting Analysis
The protein samples were spotted on a nitrocellulose membrane to assess Lf accumulation. The membrane was blocked with 1% skim milk, then washed in PBS and shaken for 2 h. After 1 h of incubation with the primary antibody (Rabbit Anti Histag), the nitrocellulose membrane was washed 3 times for 5 min each with sufficient PBS-T buffer. The membrane was then incubated in an alkaline phosphatase-secondary antibody conjugate (Anti Rabbit IgG + AP) at an appropriate dilution (1:30,000) in PBS buffer for 1 h. The rbLf was detected by incubating the membrane in the substrate mixture of NBT and BCIP in AP buffer for 10–30 min until color development. The color reaction was stopped by distilled water. The expression level was determined via dot-blot by densitometry in a Gel Doc system.
3. RESULTS AND DISCUSSION
3.1. Batch Fermentation in the 2-L Bioreactor
Heterozygous protein expression in shake cultures of P. pastoris is usually performed in a BMMY medium; however, its composition is not optimized in specific cases and the cost is high; therefore, modifications of the culture medium have been proposed for large-scale application [22]. Our previous studies investigated the alternative optimized medium and conditions for the expression of bLf in P. pastoris KM71H-3 in a 250-mL conical flask. Several batch cultures were performed to optimize medium components [21]. The optimal culture medium was determined as 1.5% (NH4)2 SO4, 1.7% KH2PO4, 0.2% MgSO4.7H2O, and 0.25% corn-steep, 0.1M phosphate buffer, pH 6.0. The fermentation parameters included an initial OD600 of 0.5, a temperature of 28°C, a stirring speed of 250 rpm, and the addition of 0.5% methanol to the medium after every 24 h of fermentation. Under these conditions, the maximum biomass was reached after 96 h of fermentation, corresponding to an OD600 value of 53.80, equivalent to 10.66 g/L of dry cell weight [Figure 1].
 | Figure 1: The growth curve of P. pastoris KM71H-3 in the conical flask. [Click here to view] |
In this study, batch fermentation was conducted in a 2-L bioreactor with the same medium as the flask culture to determine the influence of the post-optimized medium on the bioreactor under controlled conditions of pH, agitation, and oxygen concentration. The fermentation kinetics are shown in Figure 2. The pO2 was approximately maintained at 50% in the first 4 h, corresponding to the lag phase of the yeast strain P. pastoris KM71H-3, then it decreased sharply and reached the control pO2 level of 30% after 6 h of fermentation, corresponding to the log phase. To maintain the pO2, the agitation was automatically increased from 200 to 702.8 rpm at 17 h. The yeast strain entered the stationary phase at 36 h and lasted for around 108 h. The maximum OD600 achieved at 73.90 after 96 h of fermentation, corresponding to a dry cell weight of 14.68 g/L [Figure 2].
 | Figure 2: Kinetic of the batch fermentation of P. pastoris KM71H-3 in the 2-L bioreactor. (a) Parameters of fermentation process; and (b) time course of growth curve and dry cell weight. [Click here to view] |
A comparision of the growth curves of strain P. pastoris KM71H-3 in a conical flask and a 2-L bioreactor revealed that the stages of growth were similar. However, each phase had a distinct growth rate. Specifically, the cell density attained in the bioreactor was greater than that in the conical flask. The highest OD600 value in the bioreactor was 1.37 times higher than that in the flask. This result can be explained by the fact that the pH and pO2 were maintained at constant levels of 6.0 and 30% in the bioreactor, which were more suitable conditions for the growth and development of the yeast strain P. pastoris KM71H compared to uncontrolled conditions in the flask.
Despite batch fermentation in a 2-L bioreactor, the highest biomass of P. pastoris grew 1.37 folds in comparison to that in the conical flask. However, compared to earlier reports, this biomass value was significantly lower. With regard to the fed-batch fermentation of the P. pastoris KM71 strain in a 10-L bioreactor, Li et al. reported a maximum OD600 of 200. In this work, gradient glycerol solution feeding with DO control at 30–35% was applied as a combination feeding strategy [23]. Another report by Liu et al. presented that P. pastoris GS 115 reached the highest OD600 value of 405, which corresponds to 100 g/L of DCW when using the DO-stat approach [24].
According to our observations in the previous experiments, the accumulation of target proteins from recombinant P. pastoris was usually proportional to the amount of biomass. In the next experiments, fed-batch fermentation strategies were used to increase the biomass of P. pastoris in a 2-L bioreactor, thereby increasing the amount of lactoferrin produced.
3.2. Fed-Batch Fermentation
To enhance the cell density, it is necessary to extend the log phase in the fermentation process. Fed-batch processes have been shown to be the most effective at reducing undesirable effects like catabolize repression and by-product formation [25]. Nutrient feeding was used to overcome starvation and to extend the growth of P. pastoris. In general, to avoid a long oxygen limitation in the bioreactor, which led to ethanol accumulation [26] and, as a consequence, inhibition of recombinant protein production, the growth phase was divided into two stages, including the glycerol batch phase and the glycerol fed-batch phase. It is clear that feeding strategies are needed for the high-density growth of P. pastoris.
In the previous researches in fed-batch fermentation of P. pastoris strains [12,13,26] glycerol was supplied in two stages: Batch and fed-batch phases. At these stages, glycerol was utilized to boost cell biomass since it promotes cell development more effectively than methanol does [9,27]. The duration of glycerol feeding depends on the desired cell density before methanol induction. We found that not only glycerol acts as the carbon source for the development and growth of P. pastoris, but nitrogen is also important for the growth of this P. pastoris strain. In addition, protein synthesis also requires much nitrogen, especially lactoferrin protein [28]. Furthermore, it is probable that the nitrogen concentration in the batch medium was insufficient for the yeast strain P. pastoris to reach such high-cell densities as those previously reported. Therefore, two nutrient-feeding strategies were applied to determine suitable conditions for obtaining the highest biomass. They were fed glycerol solution (i) and nutrient solutions containing glycerol and nitrogen sources and (ii) with the ratio of carbon (glycerol) to nitrogen (NH4)2SO4 corn-steep) of 3:1.
For the first strategy, the obtained results are shown in Figure 3. The lag phase was in the first 5 h, which was similar to batch fermentation. The log phase was extended from 5 h to 33 h. The dissolved oxygen concentration gradually decreased from approximately 100% to about 20% as the strain entered the log phase, whilst the stirring speed increased from 200 to 750 rpm to maintain pO2 at 20%. The glycerol solution was added to the bioreactor after 15 h of fermentation and lasted for the next 20 h. The OD600 obtained using fed-batch fermentation with glycerol was 74.80 ± 0.09, 1.86 times higher than batch fermentation (the OD600 was only 40.1) after 35 h of fermentation, and reached the highest value of 98.05 ± 0.12 after 48 h. For the second feeding strategy with a mix-feed of glycarol and nitrogen source, the kinetics of fermentation parameters are shown in Figure 4. All parameters were well controlled. In this experiment, the highest OD600 reached 362.67 ± 2.04 after 51 h of fermentation, corresponding to a dry cell weight of 72.43 ± 0.41 g/L. The amount of biomass increased significantly. The cell density was 4.9 times higher than that of batch fermentation and 3.7 times higher than that of fed-batch fermentation by feeding only glycerol. This result can be explained by the fact that nitrogen is an indispensable source of the nutrients required to construct yeast cells. In addition, cornsteep serves as a nitrogen source and an important material with high nutritional value as well. Cornsteep contains biotin and relatively high levels of several important vitamins, such as B2, B3, B6, and B7. These components are essential for the growth and development of the yeast strain. In the glycerol feeding approach, glycerol alone cannot offer sufficient nutrition for cell division at a high-cell density. Similar results were reported by Liu et al. (2020) with an OD600 of 384 in a fed-batch fermentation of P. pastoris for the production of beta-glucosidase [24]. A shorter cultivation time was performed considering the cell stat at the end of the fermentation, which would have beneficial effects on the downstream process and the total cost of protein production.The specific growth rates of the P. pastoris KM71H-3 strain in three fermentation processes were compared [Figure 5]. It can be seen that in the two fed-batch fermentation processes, the specific growth rates (μ) were much higher than in the batch fermentation. The highest growth rates of P. pastoris KM71H-3 when fed a mixed solution of glycerol and nitrogen, and feeding glycerol solution were 0.12 (μ, 1/h) and 0.075 (μ, 1/h) only after 24 h of fermentation, respectively. In batch fermentation, the specific growth rate was much lower and decreased over time. The highest growth rate in batch fermentation was 0.064 (μ, 1/h) at 12 h, then down to 0.042 (μ, 1/h) at 24 h and dropped to 0.02 (μ, 1/h) after 48 h of fermentation. These results could be explained by the fact that the concentration of glycerol gradually decreased during batch fermentation, leading to a decrease in the specific growth rate. With fed-batch fermentation, the growth rate was maintained and even increased due to an adequate supply of carbon. Rumjantsev et al. (2014) found that nutrient feeding solutions containing both carbon and nitrogen had a higher growth rate due to the supply of sufficient carbon and nitrogen sources for cell building [28]. It should be noted that in the nitrogen and carbon nutrient solutions, we added an extra amount of corn steep; this ingredient contained a variety of sugar and mineral sources, so the results obtained from the biomass were surprisingly high.
 | Figure 3: Kinetics of the fed-batch fermentation of P. pastoris KM71H-3 with glycerol solution. (a) Fundamental parameters for fermentation process; and (b) time course of growth curve. [Click here to view] |
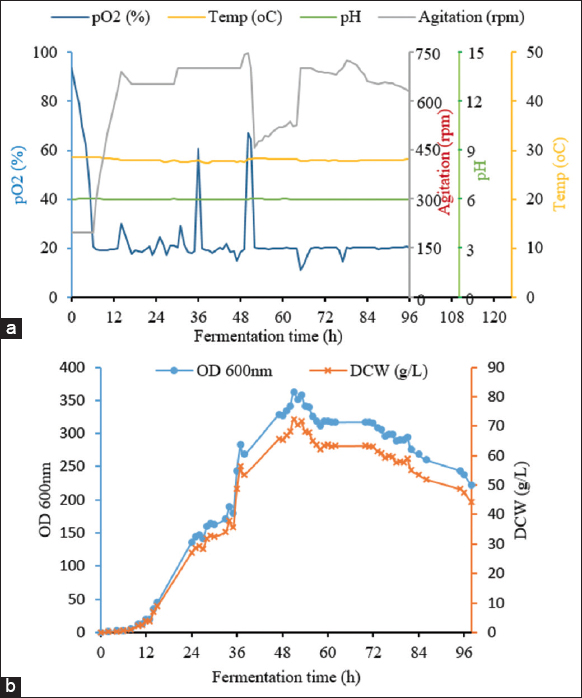 | Figure 4: Kinetics of the fed-batch fermentation of P. pastoris KM71H-3 with mix-feeds of glycerol and nitrogen solution. (a) Fundamental parameters for fermentation process; and (b) time course of growth curve. [Click here to view] |
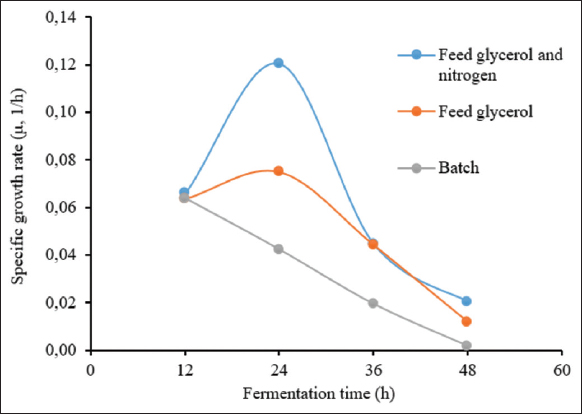 | Figure 5: Specific growth rate of P. pastoris KM71H-3 in three HCDF strategies. [Click here to view] |
3.3. Scaling-up P. pastoris KM71H-3 Fermentation in the 10-L and 100-L Bioreactors for Production of Lactoferrin
The oxygen transport coefficient model and device operating parameters were calculated using Matlab software to ensure the similarity of volumetric oxygen transport in 2-L and 10-L bioreactors. Accordingly, the stirring and aeration speed indicators were selected; specifically, the 10-L bioreactor was operated at 250 rpm–2.96 vvm and 520 rpm–1.83 vvm, respectively; while the 2-L bioreactor was operated at 400 rpm–1vvm and 700–1vvm, respectively.
For HCDF of P. pastoris in the 10-L bioreactor, the same medium and feeding conditions were used as in the 2-L bioreactor. The stirring and aeration speeds were used as calculated above. The growth curve showed similar biomass obtained from the fed-batch fermentation at the 2-L scale. In this experiment, the growth curve of the fed-batch fermentation process with feeding glycerol and nitrogen (C: N of 3:1) was used as the predicted curve for the 10-L scale. The result showed that the shapes of the growth curves of P. pastoris KM71H-3 in the 10-L bioreactors appeared to be predictions [Figure 6]. This result indicated that upgrading the fermentation scale of P. pastoris KM71H-3 from 2-L to 10-L had been successful. The biomass was obtained at a high level and reached the highest OD600 of 338.20 ± 3.38, corresponding 67.54 ± 0.68 g/L to after 49 h of fermentation.
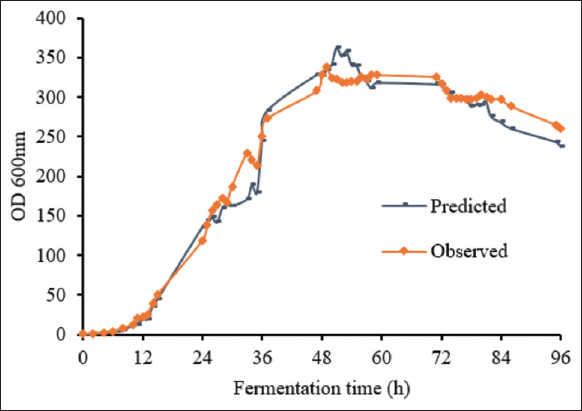 | Figure 6: The prediction and observation of the growth curves of P. pastoris KM71H-3 in fed-batch fermentation 10-L bioreactor. The time course of the growth curve of P. pastoris KM71H-3 in the 2-L bioreactor was used as the predicted curve in the 10-L bioreactor. [Click here to view] |
Scaling up the fermentation process of P. pastoris KM71H-3 from 10-L to 100-L bioreactor was conducted in the same manner as scaling up from a 2-L to a 10-L bioreactor. The medium and feeding strategies were similar to those used on the 10-L scale. The results indicated that after 43 h of fermentation, the highest OD600 value reached 375.50 ± 2.98, remained this level for up to 72 h, and then decreased to 340 ± 3.45 after 93 h of fermentation [Figure 7a]. The highest OD 600 and dry cell weight achieved at 100L scale was 375.50 ± 2.98 and 75.00 ± 0.60 g/L, respectively. Similar results were observed for lactoferrin production during fermentation on a 100-L bioreactor compared to smaller 2-L and 10-L bioreactors [Figure 7b].
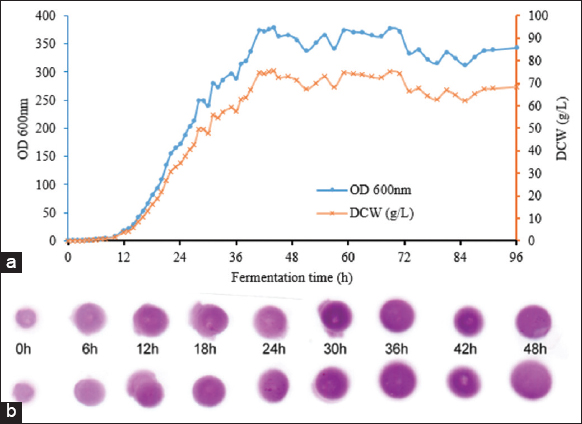 | Figure 7: Fed-batch high-cell density fermentation of P. pastoris KM71H-3 in the 100-L bioreactor. (a) Growth curve (b) time course of Dot-blot results of intra-cellular protein (lactoferrin, 0h is the starting time of induction). [Click here to view] |
The scaling-up of the microbial fermentation process is an essential step that bridges laboratory research and commercialization. For the P. pastoris system, although significant attention has been paid to optimizing HCDF, including DO levels and methanol feeding strategies, there have been only a few publications that focus on scaling-up, such as Liu et al. (2016), who developed a process for HCDF of P. pastoris for the production of glycoside hydrolase LXYL–P1–2 in the 1000-L bioreactor [22]. To our knowledge, no publication on the large-scale production of recombinant lactoferrin has been reported.
3.4. Methanol Induction Strategy for Expressing bLf in P. pastoris KM71H-3
Methanol concentration is a critical parameter in P. pastoris cultivation since it influences both growth and heterologous gene expression under the AOX promoter. Our previous study reported that methanol was induced at a suitable concentration of 0.5% for each 24 h in the conical flask [21]. In this study, we compared the methanol induction strategies in the 2-L bioreactor: 1-time induction at the rate of 0.5% MeOH/24 h and continuous induction, ensuring the corresponding MeOH concentration of 0.5% for 24 h.
To test the efficiency of Lf accumulation at different time points after methanol induction, intracellular proteins were checked on the electrophoresis gel. The percentage of protein in the 75 kDa band on the total was considered a comparison criterion between induction conditions.
For 1-time induction, the lactoferrin protein band (75 kDa) was unclear on the electrophoresis gel. It was possible that aeration could lead to methanol leakage during fermentation to induce lactoferrin biosynthesis.
To confirm this hypothesis, we experimented with the same conditions in the bioreactor as before, but the induction process was performed by continuously adding methanol. The amount of methanol used for induction remained the same as before (methanol concentration was 0.5% every 24 h). Lactoferrin protein expression was examined by SDS-PAGE electrophoresis. The results showed that the percentage of 75 kDa protein bands increased rapidly in the first 48 h of induction, reaching 11.34%, and continued to increase slightly after 72 h and 96 h of induction, reaching 11.57% and 12.16%, respectively [Figure 8a and b]. The lactoferrin protein expression level was also checked by dot blot as shown in Figures 8c and d. The dot blot results were consistent with the results of protein electrophoresis analysis. These results suggest that the appropriate induction time was 48 h. The MeOH induction strategy should be continuous with an equivalent flow rate of 0.2 mL/min, corresponding to a MeOH concentration of 0.5% for 24 h. It fits with what has been learned before, since keeping methanol concentrations at the right level is needed to get high protein yields [10].
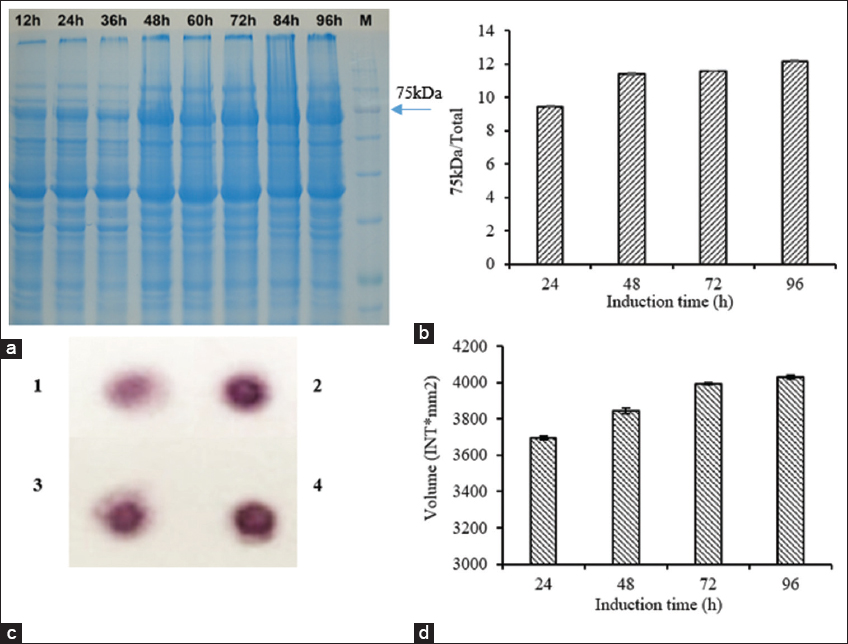 | Figure 8: Evaluation of lactoferrin expression from P. pastoris KM71H-3 with continuously feeding of MeOH strategy. (a) SDS-PAGE electrophoresis of intracellular protein; (b) quantified analysis results. The percent of rbLf (corresponding to band size 75 kDa) on the total intracellular protein (lane) of each sample was quantified by Quantity One Software. (c) Dot-blot results. Time of induction 1: 24 h; 2: 48 h; 3: 72 h; and 4: 96 h. (d) Quantified analysis results. The volume (INT*mm2) of each sample was quantified by Quantity One Software. [Click here to view] |
4. CONCLUSION
Suitable conditions for the HCDF of recombinant P. pastoris KM71H-3 strain were successfully explored in the 2-L, 10-L, and 100-L bioreactor scale. Accordingly, the fermentation process was divided into three main phases, including the glycerol batch phase (S1), the glycerol fed-batch phase (S2), and the methanol induction phase (S3). The fermentation conditions were performed at 28°C, pH 6.0 and DO remain at 20%. The optimized feeding solutions in S2 phase was mix-feed of glycerol and nitrogen sources with the ratio of C: N equivalent of 3:1. The highest cell density was obtained at an OD600 of 362.67 ± 2.04 in 2-L bioreactor, 338.00 ± 3.38 in the 10-L bioreactor and 375.50 ± 2.98 in the 100-L bioreactor which corresponds to the DCW of 72.43 ± 0.41 g/L, 67.54 ± 0.68 g/L, and 75.00 ± 0.60 g/L, resspectively. In the S3 phase, the MeOH solution was fed continuously with the corresponding concentration of 0.5% for 24 h for better recombinant lactoferrin synthesis efficiency than the 1-time induction. The results showed that it would be possible to make recombinant bLf on a large scale, which would be a key step toward making pharmaceutical intermediates more industrialized.
5. ACKNOWLEDMENTS
The authors would like to thank Assoc. Prof. Vu Nguyen Thanh from Food Industries Research Institute, Vietnam for your support.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. FUNDING
This work was supported by the research project under Grant No. DT.01.18/CNSHCB, funded by Ministry of Industry and Trade, Vietnam.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Adlerova L, Bartoskova A, Faldyna M. Lactoferrin:A review. Vet Med Praha 2008;53:457-68. [CrossRef]
2. Garcia-Montoya IA, Cendon TS, Arevalo-Gallegos S, Rascon-Cruz Q. Lactoferrin a multiple bioactive protein:An overview. Biochim Biophys Acta 2012;1820:226-36. [CrossRef]
3. Vorland LH. Lactoferrin:A multifunctional glycoprotein. APMIS 1999;107:971-81. [CrossRef]
4. Cornish J, Palmano K, Callon KE, Watson M, Lin JM, Valenti P, et al. Lactoferrin and bone;structure-activity relationships. Biochem Cell Biol 2006;84:297-302. [CrossRef]
5. Choi BK, Actor JK, Rios S, d'Anjou M, Stadheim TA, Warburton S, et al. Recombinant human lactoferrin expressed in glycoengineered Pichia pastoris:Effect of terminal N-acetylneuraminic acid on in vitro secondary humoral immune response. Glycoconj J 2008;25:581-93. [CrossRef]
6. Parkar DR, Jadhav RN, Pimpliskar MR. Antibacterial activity of lactoferrin:A review. Int J Pharm Pharm Sci 2015;4:119-27.
7. Huang N, Bethell D, Card C, Cornish J, Marchbank T, Wyatt D, et al. Bioactive recombinant human lactoferrin, derived from rice, stimulates mammalian cell growth. In Vitro Cell Dev Biol Anim 2008;44:464-71. [CrossRef]
8. Gonzalez-Chavez SA, Arevalo-Gallegos S, Rascon-Cruz Q. Lactoferrin:Structure, function and applications. Int J Antimicrob Agents 2009;33:301, e1-8. [CrossRef]
9. Liu W, Zhu P. Demonstration-scale high-cell-density fermentation of Pichia pastoris. Methods Mol Biol 2018;1674:109-16. [CrossRef]
10. Yang Z, Zhang Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris:A review. Biotechnol Adv 2018;36:182-95. [CrossRef]
11. Gmeiner C, Saadati A, Maresch D, Krasteva S, Frank M, Altmann F, et al. Development of a fed-batch process for a recombinant Pichia pastoris delta OCH1 strain expressing a plant peroxidase. Microb Cell Fact 2015;14:1. [CrossRef]
12. Liu W, Inwood S, Gong T, Sharma A, Yu LY, Zhu P. Fed-batch high-cell-density fermentation strategies for Pichia pastoris growth and production. Crit Rev Biotechnol 2019;39:258-71. [CrossRef]
13. Rodrigues D, Pillaca-Pullo O, Torres-Obreque K, Flores-Santos J, Sánchez-Moguel I, Pimenta MV, et al. Fed-batch production of Saccharomyces cerevisiae L-Asparaginase II by recombinant Pichia pastoris MUTs strain. Front Bioeng Biotechnol 2019;7:16. [CrossRef]
14. Hasslacher M, Schall M, Hayn M, Bona R, Rumbold K, Lückl J, et al. High-level intracellular expression of hydroxynitrile lyase from the tropical rubber tree Hevea brasiliensisin microbial hosts. Protein Expr Purif 1997;11:61-71. [CrossRef]
15. Werten MW, van den Bosch TJ, Wind RD, Mooibroek H, de Wolf FA. High-yield secretion of recombinant gelatins by Pichia pastoris. Yeast 1999;15:1087-96. [CrossRef]
16. Mattanovich M, Russmayer H, Scharl-Hirsch T, Puxbaum V, Burgard J, Mattanovich D, et al. Metabolomics of Pichia pastoris:Impact of buffering conditions on the kinetics and nature of metabolite loss during quenching. FEMS Yeast Res 2017;17:1-10. [CrossRef]
17. Wang M, Sun Z, Yu T, Ding F, Li L, Wang X, et al. Large-scale production of recombinant human lactoferrin from high-expression, marker-free transgenic cloned cows. Sci Rep 2017;7:10733. [CrossRef]
18. Iglesias-Figueroa B, Valdiviezo-Godina N, Siqueiros-Cendón T, Sinagawa-García S, Arévalo-Gallegos S, Rascón-Cruz Q. High-level expression of recombinant bovine lactoferrin in Pichia pastoris with antimicrobial activity. Int J Mol Sci 2016;17:902. [CrossRef]
19. Alamdari E, Niazi A, Yarizade A, Moghadam A, Aram F. Expression of a recombinant therapeutic protein, lactoferrin, in Pichiapinktm:A powerful antimicrobial protein. Biol Forum 2016;8:471-8.
20. Thuy TT, Tinh NT, Phong TQ. Research on Developing the Expression Structure of Recombinant pPICZaA:Blfopt for Bovine Lactoferrin Expression in Pichia pastoris. In:Proceedings of National Conference on Biotechnology for Sustainable Development;2018. p. 306-13.
21. Thuy TT, Thuy NT, Anh PT, Trang TT, Phong TQ. Evaluation of efficency of alternative media for production of lactoferrin from recombinant Pichia pastoris KM71H strain. In:Proceedings of National Conference on Biotechnology for Sustainable Development;2020. p. 463-9.
22. Liu W, Gong T, Wang Q, Liang X, Chen J, Zhu P. Scaling-up fermentation of Pichia pastoris to demonstration-scale using new methanol-feeding strategy and increased air pressure instead of pure oxygen supplement. Sci Rep 2016;6:18439. [CrossRef]
23. Li X, He X, Li Z, Wang F. Combined strategies for improving the production of recombinant Rhizopus oryzae lipase in Pichia pastoris. Bio Res 2013;8:2867-80. [CrossRef]
24. Liu W, Xiang H, Zhang T, Pang X, Su J, Liu H, et al. Development of a new high-cell density fermentation strategy for enhanced production of a fungus beta-glucosidase in Pichia pastoris. Front Microbiol 2020;11:1988. [CrossRef]
25. Tabandeh F, Moghaddam HR, Yakhchali B, Shariati P, Mousavian MT, Ghasemi F. Fed-batch fermentation of Bacillus clausii for efficient production of alkaline protease using different feeding strategies. Chem Eng Commun 2011;198:1063-74. [CrossRef]
26. Looser V, Bruhlmann B, Bumbak F, Stenger C, Costa M, Camattari A, et al. Cultivation strategies to enhance productivity of Pichia pastoris:A review. Biotechnol Adv 2015;33:1177-93. [CrossRef]
27. Potvin G, Zhang Z, Defela A, Lam H. Screening of alternative carbon sources for recombinant protein production in Pichia pastoris. Int J Chem React Eng 2015;14:251-7. [CrossRef]
28. Rumjantsev AM, Bondareva OV, Padkina MV, Sambuk EV. Effect of nitrogen source and inorganic phosphate concentration on methanol utilization and PEX genes expression in Pichia pastoris. ScientificWorldJournal 2014;2014:743615. [CrossRef]