1. INTRODUCTION
Polycystic ovarian syndrome (PCOS) is a common endocrine disorder among reproductive women accounting for about 10% of the female population [1,2]. About 84% of PCOS women have irregular menses and 70–80% are infertile, making it a severe disorder among reproductive women. Rotterdam consensus workshop proposed ovarian dysfunction, hyperandrogenism, and polycystic ovaries as a few critical features for identifying PCOS [3,4]. Other PCOS-associated signs include menstrual regulation, obesity, insulin resistance, an elevated luteinizing hormone (LH), and abnormal follicle-stimulating hormone (FSH) level [5,6] are also observed in PCOS women. More than 98% of PCOS women show an imbalanced LH/FSH ratio from its normal 1:1 ratio, whereas most PCOS women show a high LH/FSH ratio [7-9]. LH treated theca cell shows overexpression of cytochrome P450 family 17 subfamily A (CYP17), resulting in the conversion of progesterone to androgen [2]. In women, hyperandrogenism (high androgen level) shows a high risk of developing gestational diabetes, pregnancy-induced hypertension, and preterm birth, resulting in neonatal complications.
During pregnancy, maternal hyperandrogenism is the potential source of hyperandrogenism in developing female fetuses [10,11], which has been proposed as the leading cause of PCOS post-puberty [12]. During pregnancy, hyperandrogenic females’ placenta secretes a low amount of aromatase and a high amount of 3-beta-hydroxysteroid dehydrogenase (3-b HSD1) [13], which result in low estrogen and high androgen level in developing fetus. Animal studies in rhesus monkeys and sheep have confirmed many of the characteristic features of PCOS on excess androgen exposure during fetal life [3].
Neuroendocrine abnormalities have also been observed to be involved in PCOS. LH/FSH secretion is mediated by the gonadotropin-releasing hormone (GnRH) pulsatile secretion, where fast GnRH pulse frequency (>1 pulse/h) regulates LH surge and regular pulse frequency (<1–2 pulse/2–3 h) regulates normal LH/FSH secretion. GnRH secretion is regulated by its upstream protein kisspeptin. Various in vivo and in vitro studies revealed that kisspeptin administration stimulates GnRH and LH secretion 2-fold [14,15]. A high kisspeptin level was observed in PCOS females due to an over-active Kiss1 gene expression system [15].
Nuclear receptors, DAX1 (dosage-sensitive sex reversal, adrenal hypoplasia congenital critical region on the X chromosome, gene 1) and steroidogenic factor 1 (SF1), are critical for female fetus development. DAX1 is essential for the hypothalamus-pituitary-gonadal axis, whose overexpression results in ovary development, while down expression leads to the development of the Wolffian duct, which participates in the formation of the male reproductive organ [16,17]. DAX1 mutation results in developmental abnormality, including deficient hypothalamic GnRH secretion and adrenal hypoplasia [18]. The promoter region of the DAX1 gene has two SF1 binding sites, which results in transcriptional activation [17]. SF1 protein also regulates the transcription of many other genes involved in the adrenal gland and gonad development.
An alteration in the level of steroid hormone (androgen, estrogen, progesterone, and testosterone) reportedly affects GnRH secretion and later on the development of ovarian follicles [14,15]. Nuclear receptors such as DAX-1 (dosage-sensitive sex reversal, adrenal hypoplasia congenital critical region on the X chromosome, gene 1) and SF-1also affect the level of various sex hormones and ovary formation [16,17]. Hence, in the present work, an in silico analysis was carried out to evaluate the effect of hyperandrogenicity during fetal ovarian development. The study involved evaluating the effect of these steroid hormones on the promoter region of KISS1 gene; DAX1 gene and also on the binding of kisspeptin (involved in GnRH secretion) and SF1 protein (involved in DAX1 expression modulation) using molecular docking.
2. MATERIALS AND METHODS
2.1. Molecular Modeling
There was no reported tertiary structure for kisspeptin protein (Uniprot ID: Q15726) and SF1 protein (Uniprot ID: Q13285) in the RCSB-PDB database and hence was modeled computationally. The tertiary structure of SF1 protein (Uniprot ID: Q13285) was modeled using the homology modeling tool Swiss-Model web server. No close structure with >30% sequence similarity was available for kisspeptin protein; hence, it was modeled using an Ab-initio-based modeling tool I-TASSER [19-21]. Modeled protein structures were further validated for overall structure quality using various online tools. Ramachandran plot was measured to analyze the stereochemical and overall structure quality [22]. The Q-mean score was calculated using the Swiss-Model web server for local and global analysis of modeled protein structures [23]. ProSA Z-score highlighted the overall model quality score and was calculated using ProSA server [24]. Verify 3D score was calculated for compatibility of 3D atomic model with its amino acid sequence [21]. The structures were subjected to energy minimization, to remove unfavorable non-bonded contacts, using the YASARA Energy Minimization server [25].
The promoter region of gene Kiss1 (Gene ID: 3814) and DAX1 (Gene ID: 190) was predicted using online promoter prediction servers “Neural Network promoter prediction server” [26]; “Soft berry FPROM Human promoter prediction server” [27]; and ‘Promoter 2.0 prediction server” [28]. Consensus promoter regions obtained from these servers were chosen, and tertiary structures of these promoters were modeled using the “model.it” server and energy minimization was done using the AMBER force field [29].
The 3D structures of all studied steroids, that is, androgen, estrogen, progesterone, and testosterone with PubChem CIDs 6128, 5757, 5994, and 6013, respectively, were downloaded from the NCBI PubChem database [Figure 1]. Molecular file format converter Open Babel [30] was used to convert mol2 files of steroids to PDB files. All the structure files were subjected to “dock prep” module of UCSF Chimera v1.15 [31] for docking studies.
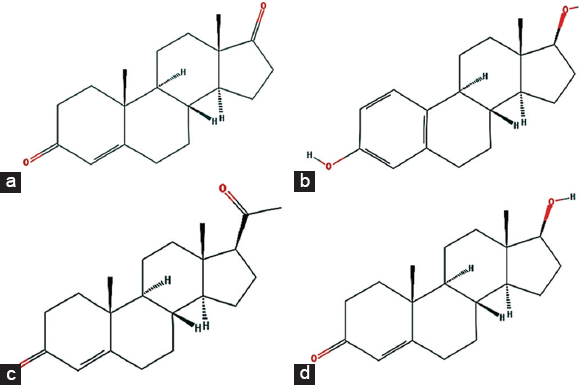 | Figure 1: Structure of studied steroid hormones – (a) androgen, (b) estrogen, (c) progesterone, and (d) testosterone. [Click here to view] |
2.2. Molecular Docking
All selected steroids (androgen, estrogen, progesterone, and testosterone) were individually docked against modeled kisspeptin protein, SF1 protein, and kiss1 gene promoter region using Autodock v4.2.6 [32]. PDB structure of kisspeptin protein and kiss1 gene promoter regions was converted to PDBQT using MGL Autodock tool v1.5.6 [32,33]. Only polar hydrogens were added, and charges (Kollman and Gasteiger) were assigned to maintain homogeneity throughout the structure. After assigning torsions and rotatable bonds, individual steroid structures were converted in PDBQT format. A grid box was generated with a default spacing value of 0.375Å. Using the genetic algorithm (GA) as a search parameter, a total of 100 independent runs with a step size of 0.2Å for translation and 5? for orientations and torsions were performed. The maximum number of gestations was set to 1000. The maximum number of top individuals that automatically survived was set to 1 with a mutation rate of 0.02, crossover rate of 0.8, cluster tolerance of 0.5Å, and external grid energy 1000.
The DNA promoter region of gene DAX1 was docked against free SF1 protein and SF1 complexed with different steroids (androgen, estrogen, progesterone, and testosterone) using a protein nucleotide dock module of Hex 8.0 [34-36]. Free SF1 protein and SF1 protein with steroid dock complexes were treated as the receptor, and the DAX1 gene promoter region was uploaded as the ligand. Using shape+electro as correlation type, 0.6 as grid dimension, 180 as receptor, and ligand range with a step size of 7.5, a total of 25 searches were performed. The docking visualization and analysis were carried out using LigPlot+ [37].
3. RESULTS AND DISCUSSION
3.1. Molecular Modeling
The 3D structure of human kisspeptin protein (Uniprot ID: Q15726) was modeled by ab-initio modeling approach using the web server I-TASSER. The modeled tertiary structure of kisspeptin [Figure 2a] was selected after structure evaluation and validation [Table 1]. Ramachandran plot [Figure 3a] showed 93.4% residue in the favored and allowed region, signifying a good model, as described in earlier report [22]. Furthermore, the ProSA Z-score for the model was observed optimal of −4.46 [Figure 4a] with a satisfactory Q-mean score of −3.22. The Verify 3D pass statement confirmed no error with experimental and theoretical models of proteins, which suggested the conformational stability of protein model, in accordance with the previous findings [21,23]. The selected structure was energy minimized using the YASARA energy minimization server to remove unfavorable non-bonded contacts in concurrence to previous finding [25], and the minimized structure was used for further study.
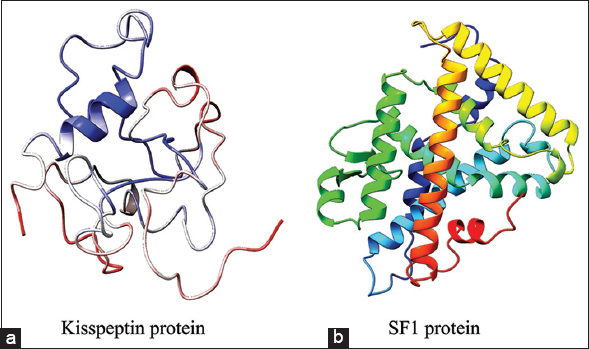 | Figure 2: The modeled tertiary structure of proteins – (a) kisspeptin, and (b) SF1 [Click here to view] |
Table 1: Structure validation data for modeled kisspeptin protein and SF1 protein.
| Validation tools | Kisspeptin protein model | SF1 protein model |
|---|---|---|
| Ramachandran plot | ||
| Favored region | 64.5% | 95.1% |
| Allowed region | 28.9% | 4.5% |
| Disallowed region | 6.5% | 0.4% |
| Ramachandran Z-score | −4.829 | 1.331 |
| Q-mean score | −3.22 | −0.96 |
| Verify 3D | Pass (89.86%) | Pass (84.02%) |
| ProSA Z-score | −4.46 | −6.69 |
The homology model of SF1 protein was generated using the “Swiss-Model” web server and “human nuclear receptor sf-1 (PDB: 4QJR1A)” as template, the later showing 99.18% sequence similarity with human SF1 protein (Uniprot ID: Q13285). The 3D structure of SF1 [Figure 2b] was selected after structure evaluation and validation [Table 1]. Ramachandran plot for SF1 showed 95.1% residue in the favored region [Figure 3b] suggesting it as a good working model, in concurrence to earlier report [22]. The ProSA Z-score [Figure 4b] of −6.69 confirmed no error with the experimental and theoretical model of proteins [24]. The Q-mean score of −0.96 showed good structure prediction compared to its template. The selected structure was energy minimized using the YASARA energy minimization server to remove unfavorable non-bonded contacts in concurrence to previous finding [25] and was used for further study.
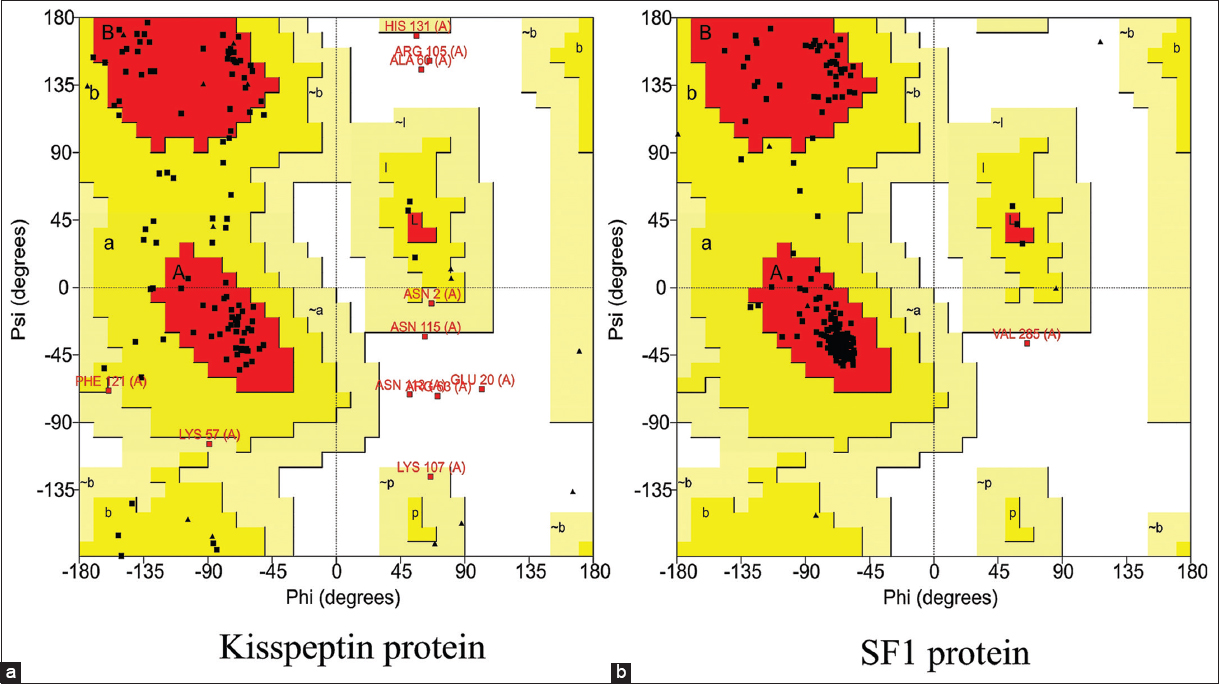 | Figure 3: Ramachandran plot for modeled protein structures of (a) kisspeptin and (b) SF1. [Click here to view] |
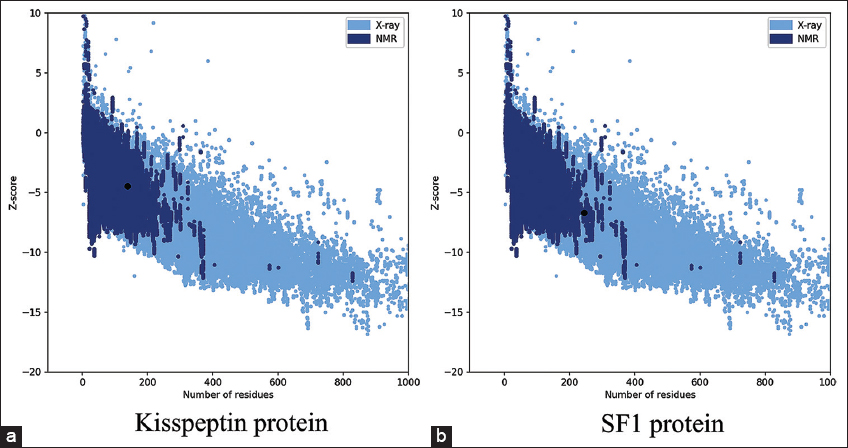 | Figure 4: ProSA model quality score graph for modeled protein structures – (a) kisspeptin (Z-score= −4.46) and (b) SF1 (Z-score= −6.69). [Click here to view] |
The DNA promoter regions for gene Kiss1 and DAX1 were identified using machine learning approaches with a promoter range of 3175–3225 and 1514–1564, respectively [Table 2]. The tertiary structures for both the gene promoter regions were predicted using the “model.it” web server with a straight B-DNA parameter setup and were used for further molecular docking studies [Figure 5].
Table 2: Promoter prediction score for genes – Kiss1 and DAX1.
| Promoter prediction servers | Kiss1 gene | DAX1 gene |
|---|---|---|
| Start | 3175 | 1514 |
| End | 3225 | 1564 |
| Promoter position | 3215 | 1549 |
| Neural network promoter prediction score | 0.90 | 0.99 |
| FPROM human promoter prediction score | 7.798 | 8.921 |
| Promoter 2.0 prediction score | 1.166 | 1.244 |
| LDF | +3.608 | +7.283 |
| Promoter sequence | CCAGTCACTCCTATATAT GGCATC | CCTGCGTGCGCGCTAGG TATAAATA |
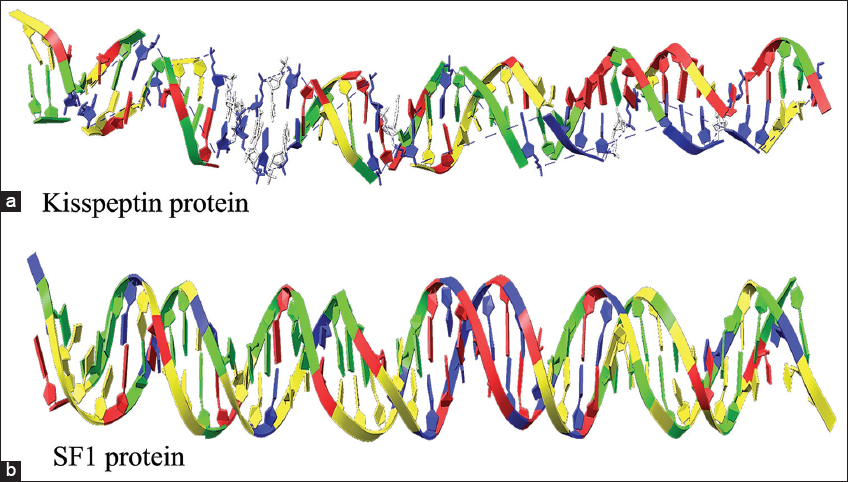 | Figure 5: The modeled tertiary structure of promoter regions of gene (a) Kiss1 and (b) DAX1. [Click here to view] |
3.2. Molecular Docking
All the steroids (androgen, estrogen, progesterone, and testosterone), taken for the present study, were docked against the promoter region of the kiss1 gene, kisspeptin protein, and SF1 protein. The binding affinities of free SF1 protein and its dock complex with (androgen, estrogen, progesterone, and testosterone) were estimated by docking interaction on promoter region of DAX1 gene. All studied proteins and promoter regions showed good binding of steroids with binding energies ranging from −7.50 to −9.70 Kcal/Mol.
The binding score for docking interaction of steroids (androgen, estrogen, progesterone, and testosterone) with the promoter region of Kiss1 gene ranged from −8.25 to −9.66 Kcal/Mol, where androgen showed minimum binding energy of −9.66 Kcal/Mol [Table 3]. Steroids androgen and progesterone had the same binding score and a common binding location toward the 5’ region over the TATA box (A [a13, t14, a15, t16}, B [a37, t38, a39]). In contrary, the steroids – estrogen and testosterone, showed similar binding score and had the same binding site with common interacting residues toward the center of the promoter region (A [t23, c24, t25], B [g27, a28, t29, g30]) [Figure 6]. The androgen and progesterone binding over TATA box may be predicted to prevent binding of TATA box binding protein which may inhibit the expression of kiss1 gene. In contrast, testosterone and estrogen bind downstream of the TATA box and do not appear to regulate Kiss1 gene expression [Figure 7]. This androgen- and progesterone-mediated Kiss1 gene regulation may affect kisspeptin signaling in the hypothalamus and downstream regulation of the hypothalamus-pituitary-gonads axis to cause an imbalanced steroid hormone level.
Table 3: Binding energy and interacting residues of Kiss1 gene promoter region when docked against studied steroid hormones (androgen, estrogen, progesterone, and testosterone).
| Steroids | Binding energy (KCal/Mol) | H-bond forming residues | Interacting residue |
|---|---|---|---|
| Androgen | −9.66 | - | A (a13, t14, a15, t16). |
| Estrogen | −8.25 | B (g27 (3.01Å)). | A (t23, c24, t25), |
| Progesterone | −9.62 | - | A (a13, t14, a15, t16), |
| Testosterone | −8.27 | - | A (t23, c24, t25), |
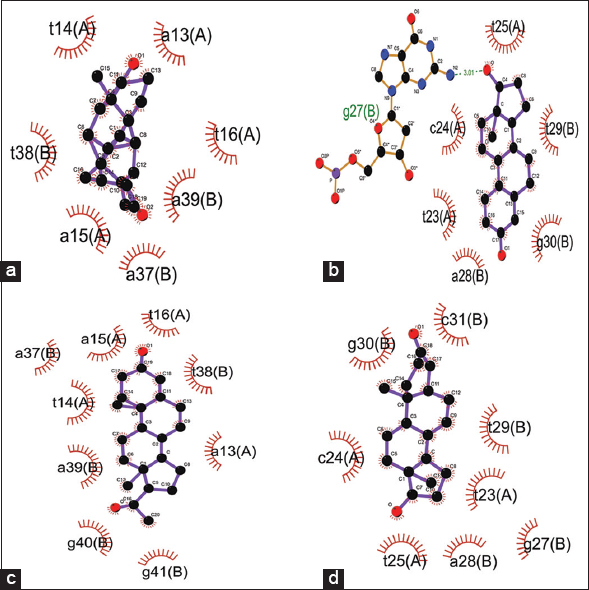 | Figure 6: Docking interactions of the KISS1 gene promoter region with steroid hormones – (a) androgen, (b) estrogen, (c) progesterone, and (d) testosterone. [Click here to view] |
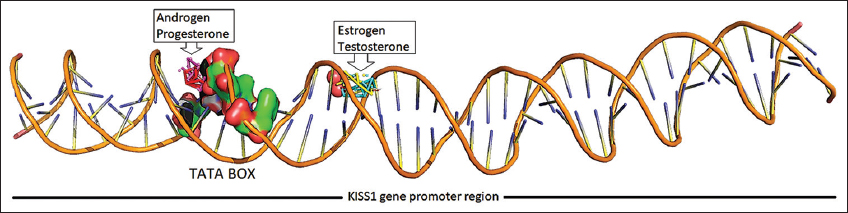 | Figure 7: Steroid hormones binding over promoter region of KISS1 gene. [Click here to view] |
The binding score of steroids (androgen, estrogen, progesterone, and testosterone) when docked against kisspeptin protein ranged from −8.80 to −9.46 Kcal/Mol. Androgen showed minimum binding energy of −9.46 Kcal/Mol, followed by testosterone, progesterone, and estrogen with a binding energy of −9.38, −9.30, and −8.80 Kcal/Mol, respectively [Table 4]. Androgen and progesterone showed the same binding pockets represented by 9-amino acids: Gln8, Leu9, Leu11, Phe12, Pro84, Leu86, Ser87, Val101, and Leu102. Estrogen and testosterone showed another common binding pocket represented by Leu44, Ala45, Pro46, Glu48, Leu51, Cys53, Glu55, Thr61, and Gly118 as common nearby residue including Phe117 and Leu119 as H-bond forming amino acids [Figure 8]. Kisspeptin protein region 112–121 (kisspeptin-10) has been observed earlier as critical for binding to its receptor (Gpr54) [38-40]. Our studied hormones (estrogen, progesterone, and testosterone) showed their binding in and around the receptor-binding region of kisspeptin protein, which may be predicted to hamper kisspeptin binding to its natural receptor. This can be due to the negative feedback mechanism of sex steroids to regulate their secretion by inhibiting kisspeptin binding to its receptor [41]. However, androgen was found to bind at different location, leaving receptor binding pocket free to trigger kisspeptin binding to its receptor. Thus, it may be inferred that in the case of hyperandrogenism, androgen outnumbered other sex steroids to regulate GnRH surge. This altered GnRH surge may further produce imbalanced LH and FSH hormones through the overstimulated hypothalamus-pituitary-gonadal axis.
Table 4: Binding energy and interacting residues of kisspeptin protein when docked against studied steroid hormones (androgen, estrogen, progesterone, and testosterone).
| Steroids | Binding energy (KCal/Mol) | H-bond forming residues | Interacting residue |
|---|---|---|---|
| Androgen | −9.46 | - | Gln8, Leu9, Leu11, Phe12, Pro84, Leu86, Ser87, Val101, and Leu102. |
| Estrogen | −8.80 | Phe117 (2.91Å), Leu119 (2.72Å). | Leu44, Ala45, Pro46, Gly47, Glu48, Leu51, Cys53, Glu55, Thr61, Phe117, Gly118, and Leu119. |
| Progesterone | −9.30 | - | Gln8, Leu9, Leu11, Phe12, Pro84, Gly85, Leu86, Ser87, Ala100, Val101, Leu102, Phe121, and Ala127. |
| Testosterone | −9.38 | Phe117 (2.88Å), Leu119 (2.74Å). | Leu44, Ala45, Pro46, Glu48, Leu51, Cys53, Glu55, Thr61, Phe117, Gly118, and Leu119. |
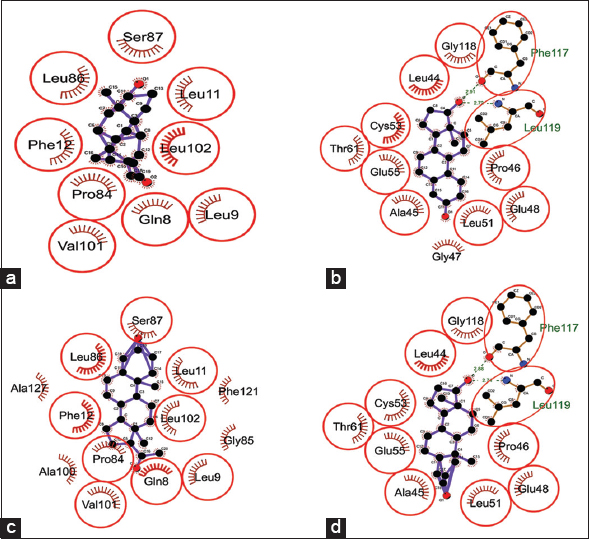 | Figure 8: Dock interactions of kisspeptin protein with steroid hormones – (a) androgen, (b) estrogen, (c) progesterone, and (d) testosterone. [Click here to view] |
The docking interactions of steroids (androgen, estrogen, progesterone, and testosterone) against SF1 protein had the binding scores in the range of −7.59–−9.51 Kcal/Mol. Progesterone showed minimum binding energy of −9.51 Kcal/Mol followed by androgen, testosterone, and estrogen with a binding energy of −8.78, −8.38, and −7.59 Kcal/Mol, respectively [Table 5]. Androgen, progesterone, and testosterone showed the same binding pocket with Leu306 and His310 as nearby amino acid residues, while estrogen showed separate binding pocket [Figure 9]. All studied sex steroids showed their binding in the leucine-rich ligand-binding domain of SF1 protein, which might disturb SF1 binding to the promoter region of the DAX1 gene, as reported earlier [42,43]. The high binding affinity of progesterone, androgen, and testosterone toward SF1 protein binding can be predicted to be crucial for expressing other gonadal developmental genes.
Table 5: Binding energy and interacting residues of SF1 protein when docked against studied steroid hormones (androgen, estrogen, progesterone, and testosterone).
| Steroids | Binding energy (KCal/Mol) | H-bond forming residues | Interacting residue |
|---|---|---|---|
| Androgen | −8.78 | - | Ser303, Leu306, Val307, His310, Leu347, Ala351, Leu429, Ala433, and Leu437. |
| Estrogen | −7.59 | Met446 (2.70Å), Glu454 (3.08Å). | His441, Asn444, Met446, Pro447, Arg448, Ans449, Asn450, Ile453, and Glu454. |
| Progesterone | −9.51 | - | Trp302, Ser303, Leu306, Val307, His310, Leu344, Leu347, Val348, Ala351, Ala433, Lys434, and Leu437. |
| Testosterone | −8.38 | Leu306 (2.80Å), Asp309 (2.98Å), Tyr436 (3.01Å). | Leu265, Met268, Ala269, Thr272, Leu306, Asp309, His310, Val326, and Tyr436. |
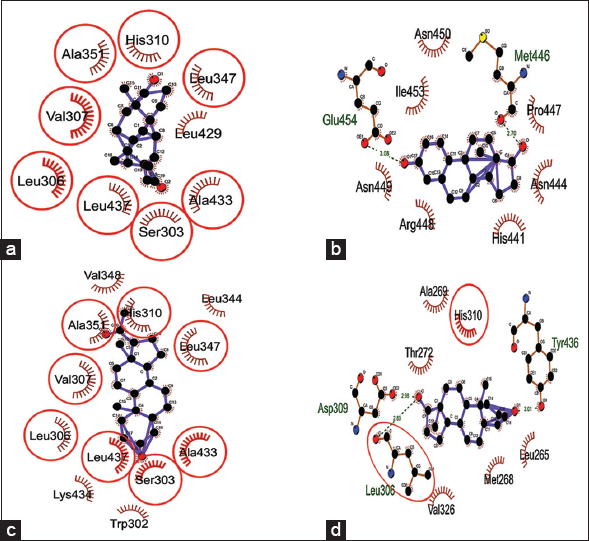 | Figure 9: Dock interactions of SF1 protein with steroid hormones – (a) androgen, (b) estrogen, (c) progesterone, and (d) testosterone. [Click here to view] |
Docking analysis of SF1 protein and SF1 protein complexed with individual steroid (androgen, estrogen, progesterone, and testosterone) against the promoter region of DAX1 gene was done using Hex 8.0. The unbound SF1 protein showed a comparative minimum binding score of −1299.84 against the DAX1 gene promoter region compared to SF1 protein bound with steroid [Table 6]. SF1 protein showed two different binding positions when docked against the DAX1 gene promoter region. The unbound SF1 protein and SF1 protein bound to progesterone showed binding at 5’ upstream region near TATA box whereas the other three studied hormones bound to SF1 protein showed their binding interactions at 3’ region of promoter region, near the transcription start site [Figure 10a-e]. DAX1 gene promoter region has 2 SF1 binding sites for the expression of the DAX1 gene [17]. Dax1 is crucial for expressing oogenesis regulated genes, steroidal differentiation-related genes, and sex differentiation [44,45]. The unbound SF1 protein showed maximum binding affinity near the TATA box of DAX1 gene, reflecting possible overexpression of the DAX1 gene in its presence, further stimulating other genes (StAR, Cyp1, and Cyp19) expression and development of the normal ovary. Earlier studies have showed that DAX1 mutant mice show a high expression of steroidogenic genes, including StAR, P450c17, P450scc, and 3β-HSD [18,46], which trigger testosterone synthesis and disturb normal oogenesis in females. The SF1 protein when complexed with studied steroids showed significantly less binding affinity [Table 6], thus hinting toward possible downregulation of DAX1 gene expression.
Table 6: Binding energy and interacting residues of the promoter region of DAX1 gene when docked against unbound SF1 protein and SF1 protein bound to studied steroid hormones (androgen, estrogen, progesterone, and testosterone).
| Molecules | Binding score | H-bond forming residues | Interacting residues |
|---|---|---|---|
| SF1 | −1299.84 | Thr252-B (a44), | A (c2, t3, g4, c5, g6, t7, g8, c9, g10, c11, g12). |
| SF1_A | −629.41 | Leu245-A (c42), | A (c41, c42, a43, c44, g46, g47, g48). |
| SF1_E | −688.26 | Gln339-A (g40), | A (g40, c41, c42, a43, c44, t45, g47). |
| SF1_P | −656.68 | Cys247-B (c43), | A (t3, g4, g6, t7, g8, c9, g10, c11, g12). |
| SF1_T | −376.35 | Thr252-B (g9), | A (g40, c41, c42, a43, c44, t45, g46, g47). |
SF1: Unbound SF1 protein; SF1_A: SF1 protein bound to androgen; SF1_E: SF1 protein bound to estrogen; SF1_P: SF1 protein bound to progesterone; SF1_T: SF1 protein bound to testosterone
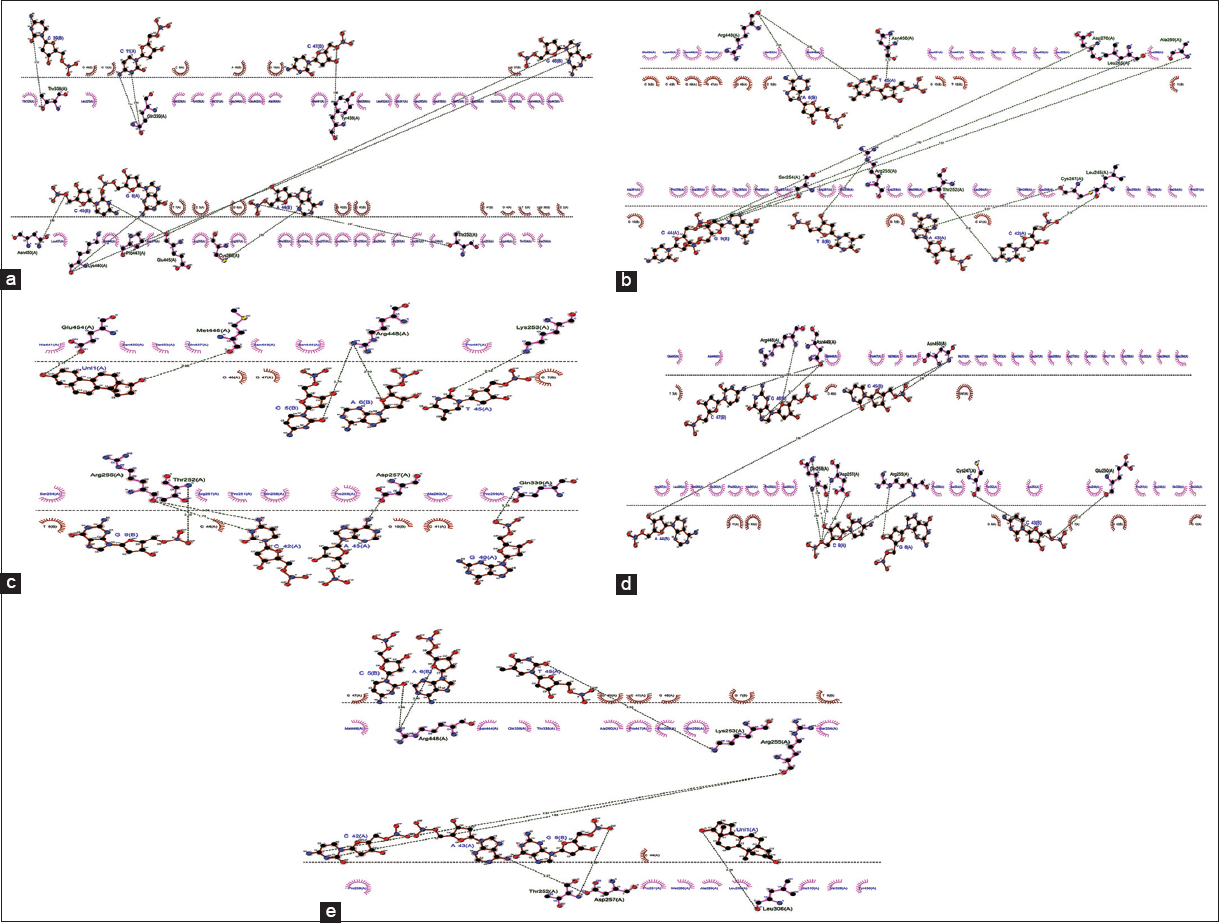 | Figure 10: Dock interaction of KISS1 gene promoter region with a) SF1 protein, b) SF1 protein dock complex with androgen, c) SF1 protein dock complex with estrogen, d) SF1 protein dock complex with progesterone, and e) SF1 protein dock complex with testosterone. [Click here to view] |
4. CONCLUSION
PCOS is a multifactorial disorder, but hyperandrogenism is the significant cause. The observations of present in silico investigation reflect that the binding of steroid hormones with SF1 protein lowers the expression of DAX1 gene. The SF1 protein is essentially required for DAX1 gene expression, and the binding of studied steroid hormones with SF1 protein may result in downregulation of DAX1 gene expression, leading to abnormal ovary development in a female fetus as well as abnormal sex steroids level. Thus, it may be concluded that over-secretion of steroid hormones (androgen, estrogen, progesterone, and testosterone) possibly affects female fetus development and hypothalamus-pituitary-gonads axis to trigger PCOS-like symptoms.
5. ACKNOWLEDGMENT
The authors acknowledge the laboratory facility provided by DBT-Bioinformatics Infrastructure Facility, Centre for Bioinformatics, M. D. University, Rohtak. PK acknowledges the SRF provided by CSIR, New Delhi.
6. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
9. FUNDING
There is no funding to report.
10. DATA AVAILABILITY
All data generated and analyzed are included within this research article.
11. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Coutinho EA, Kauffman AS. The role of the brain in the pathogenesis and physiology of polycystic ovary syndrome (PCOS). Med Sci (Basel) 2019;7:84. [CrossRef]
2. Cadagan D, Khan R, Amer S. Thecal cell sensitivity to luteinizing hormone and insulin in polycystic ovarian syndrome. Reprod Biol 2016;16:53-60. [CrossRef]
3. Franks S. Controversy in clinical endocrinology:Diagnosis of polycystic ovarian syndrome:In defense of the Rotterdam criteria. J Clin Endocrinol Metab 2006;91:786-9. [CrossRef]
4. Fauser BC. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril 2004;81:19-25. [CrossRef]
5. Witchel SF, Oberfield SE, Peña AS. Polycystic ovary syndrome:Pathophysiology, presentation, and treatment with emphasis on adolescent girls. J Endocr Soc 2019;3:1545-73. [CrossRef]
6. Yen SS, Vela P, Rankin J. Inappropriate secretion of follicle-stimulating hormone and luteinizing hormone in polycystic ovarian disease. J Clin Endocrinol Metab 1970;30:435-42. [CrossRef]
7. Coyle C, Campbell RE. Pathological pulses in PCOS. Mol Cell Endocrinol 2019;498:110561. [CrossRef]
8. Esparza LA, Schafer D, Ho BS, Thackray VG, Kauffman AS. Hyperactive LH pulses and elevated kisspeptin and NKB gene expression in the arcuate nucleus of a PCOS mouse model. Endocrinology 2020;161:bqaa018. [CrossRef]
9. Malini NA, Roy George K. Evaluation of different ranges of LH:FSH ratios in polycystic ovarian syndrome (PCOS) - Clinical based case control study. Gen Comp Endocrinol 2018;260:51-7. [CrossRef]
10. Abbott DH, Tarantal AF, Dumesic DA. Fetal, infant, adolescent and adult phenotypes of polycystic ovary syndrome in prenatally androgenized female rhesus monkeys. Am J Primatol 2009;71:776-84. [CrossRef]
11. Sir-Petermann T, Maliqueo M, Angel B, Lara HE, Pérez-Bravo F, Recabarren SE. Maternal serum androgens in pregnant women with polycystic ovarian syndrome:Possible implications in prenatal androgenization. Hum Reprod 2002;17:2573-9. [CrossRef]
12. Homburg R. Androgen circle of polycystic ovary syndrome. Hum Reprod 2009;24:1548-55. [CrossRef]
13. Sun M, Maliqueo M, Benrick A, Johansson J, Shao R, Hou L, et al. Maternal androgen excess reduces placental and fetal weights, increases placental steroidogenesis, and leads to long-term health effects in their female offspring. Am J Physiol Endocrinol Metab 2012;303:E1373-85. [CrossRef]
14. Novaira HJ, Ng Y, Wolfe A, Radovick S. Kisspeptin increases GnRH mRNA expression and secretion in GnRH secreting neuronal cell lines. Mol Cell Endocrinol 2009;311:126-34. [CrossRef]
15. Tang R, Ding X, Zhu J. Kisspeptin and polycystic ovary syndrome. Front Endocrinol (Lausanne) 2019;10:298. [CrossRef]
16. Calvari V, Alpigiani MG, Poggi E, Podesta B, Camerino G, Lorini R. X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism:Report on new mutation of the DAX-1 gene in two siblings. J Endocrinol Invest 2006;29:41-7. [CrossRef]
17. Kelly VR, Hammer GD. LRH-1 and Nanog regulate Dax1 transcription in mouse embryonic stem cells. Mol Cell Endocrinol 2011;332:116-24. [CrossRef]
18. Caron P, Imbeaud S, Bennet A, Plantavid M, Camerino G, Rochiccioli P. Combined hypothalamic-pituitary-gonadal defect in a hypogonadic man with a novel mutation in the DAX-1 gene. J Clin Endocrinol Metab 1999;84:3563-9. [CrossRef]
19. Yang J, Zhang Y. Protein structure and function prediction using I-TASSER. Curr Protoc Bioinformatics 2015;52:5.8.1-15. [CrossRef]
20. Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 2008;9:40. [CrossRef]
21. Eisenberg D, Lüthy R, Bowie JU. VERIFY3D:Assessment of protein models with three-dimensional profiles. Methods Enzymol 1997;277:396-404. [CrossRef]
22. Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Calpha geometry:phi,psi and Cbeta deviation. Proteins 2003;50:437-50. [CrossRef]
23. Benkert P, Tosatto SC, Schomburg D. QMEAN:A comprehensive scoring function for model quality assessment. Proteins 2008;71:261-77. [CrossRef]
24. Wiederstein M, Sippl MJ. ProSA-web:Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007;35:W407-10. [CrossRef]
25. Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, et al. Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling:Four approaches that performed well in CASP8. Proteins 2009;77 Suppl 9:114-22. [CrossRef]
26. Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 2001;26:51-6. [CrossRef]
27. Solovyev VV, Shahmuradov IA, Salamov AA. Identification of promoter regions and regulatory sites. In:Methods in Molecular Biology. Vol. 674. Clifton, N.J.:Springer;2010. 57-83. [CrossRef]
28. Knudsen S. Promoter2.0:For the recognition of PolII promoter sequences. Bioinformatics 1999;15:356-61. [CrossRef]
29. Munteanu MG, Vlahovicek K, Parthasarathy S, Simon I, Pongor S. Rod models of DNA:Sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem Sci 1998;23:341-7. [CrossRef]
30. O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel:An open chemical toolbox. J Cheminform 2011;3:33. [CrossRef]
31. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605-12. [CrossRef]
32. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4:Automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785-91. [CrossRef]
33. Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 2016;11:905-19. [CrossRef]
34. Ritchie DW, Venkatraman V. Ultra-fast FFT protein docking on graphics processors. Bioinformatics 2010;26:2398-405. [CrossRef]
35. Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. HexServer:An FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 2010;38:W445-9. [CrossRef]
36. Ghoorah AW, Devignes MD, Smaïl-Tabbone M, Ritchie DW. Protein docking using case-based reasoning. Proteins 2013;81:2150-8. [CrossRef]
37. Laskowski RA, Swindells MB. LigPlot+:Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 2011;51:2778-86. [CrossRef]
38. Rather MA, Bhat IA, Gireesh-Babu P, Chaudhari A, Sundaray JK, Sharma R. Molecular characterization of kisspeptin gene and effect of nano-encapsulted kisspeptin-10 on reproductive maturation in Catla catla. Domest Anim Endocrinol 2016;56:36-47. [CrossRef]
39. Son HE, Kim KM, Kim EJ, Jang WG. Kisspeptin-10 (KP-10) stimulates osteoblast differentiation through GPR54-mediated regulation of BMP2 expression and activation. Sci Rep 2018;8:2134. [CrossRef]
40. Li Y, Cao Y, Wang J, Fu S, Cheng J, Ma L, et al. Kp-10 promotes bovine mammary epithelial cell proliferation by activating GPR54 and its downstream signaling pathways. J Cell Physiol 2020;235:4481-93. [CrossRef]
41. Adams C, Stroberg W, DeFazio RA, Schnell S, Moenter SM. Gonadotropin-releasing hormone (GnRH) neuron excitability is regulated by estradiol feedback and kisspeptin. J Neurosci 2018;38:1249-63. [CrossRef]
42. Robevska G, van den Bergen JA, Ohnesorg T, Eggers S, Hanna C, Hersmus R, et al. Functional characterization of novel NR5A1 variants reveals multiple complex roles in disorders of sex development. Hum Mutat 2018;39:124-39. [CrossRef]
43. Mitsis T, Papageorgiou L, Efthimiadou A, Bacopoulou F, Vlachakis D, Chrousos GP, et al. A comprehensive structural and functional analysis of the ligand binding domain of the nuclear receptor superfamily reveals highly conserved signaling motifs and two distinct canonical forms through evolution. World Acad Sci J 2019;1:264-74. [CrossRef]
44. Albrecht KH, Eicher EM. Sex determination, mouse. In:Brenner S, Miller JH, editors. Encyclopedia of Genetics. Academic Press;2001, 1816-9. [CrossRef]
45. Bizzarri C, Cappa M. Ontogeny of hypothalamus-pituitary gonadal axis and minipuberty:An ongoing debate?Front Endocrinol (Lausanne) 2020;11:187. [CrossRef]
46. Kumar S, Kim HJ, Lee CH, Choi HS, Lee K. Leydig cell-specific DAX1-deleted mice has higher testosterone level in the testis during pubertal development. Reprod Sci 2022;29:955-62. [CrossRef]