1. INTRODUCTION
Conjunctivitis is a common type of eye infection usually characterized by inflammation of conjunctivae with swelling of the conjunctival tissue, engorgement of the blood vessels, eye discharge, and irritation. Globally, conjunctivitis affects multiple subjects, and it is one of the important causes of office visits to general medical and ophthalmological clinics [1]. The commonest types of bacteria that cause bacterial conjunctivitis include Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, Klebsiella spp., Streptococcus pneumoniae, Haemophilus influenzae, Proteus mirabilis, and Neisseria gonorrhoeae [2]. Haemophilus influenzae is the causative agent and the commonest type of bacteria isolated from patients suffering from bacterial conjunctivitis [3]. It is a Gram-negative coccobacilli bacterium, and the major virulence factor is the polysaccharide capsule. Depending on the presence or absence of the capsule, H. influenzae strains can be subdivided into encapsulated (typeable) and nonencapsulated (nontypeable). Encapsulated strains were divided into six serotypes, ranging from A to F, and comparatively homologous irrespective of their serotype, while the nontypeable strains are genetically diverse [4]. It is a respiratory tract opportunistic pathogen and the largest contributor to invasive disease of Haemophilus. During the process of pathogenesis, adhesion plays a major role. Also, they may be diverse in structures like trimeric autotransporters “Hmw1, Hmw2, Hia, and Hap,” surface proteins, “outer membrane proteins,” including OMPs 1,2, 4, 5, and 6, proteins E, and proteins F, as well as surface protrusions “T4SS and HIF” and the immunogenicity and potential vaccine candidates [5]. Additionally, H. influenzae can be classified into three major categories, including (nonencapsulated, capsulated nontype b, and encapsulated type b), where type b is considered as the most virulent form [6].
The objectives of the current study are to isolate and identify H. influenzae from eye swabs of patients with conjunctivitis by the traditional culture methods, Satellitism test using “X and V” factors, and PCR technique. In addition to differentiating both encapsulated (typeable) and nonencapsulated (nontypeable) H. influenzae using bexA and bexB gene primers by the PCR assay.
2. MATERIALS AND METHODS
2.1. Eye Swabs Collection and Bacterial Identification
The eye swabs were obtained from the conjunctiva of 150 patients with conjunctivitis of both sexes admitted to Ophthalmic Unit of Hilla Teaching Hospital during the period from July to December 2020 in Hilla Province, Babylon Governorate, Iraq. The specimens were transported using specific transport media, and in the laboratory, these swabs were cultured onto the surface of blood agar, MacConkey agar, and chocolate agar. After that, the identified bacterial colonies were subjected to Gram’s stain, and specific biochemical tests such as “catalase, oxidase, Voges–Proskauer, indole, urease, nitrate reduction, and carbohydrate fermentation” were performed to reach the final identification [7].
2.2. Satellitism Test of H. influenzae
This test, used to determine H. influenzae isolates, was conducted by mixing a loopful of suspected bacterial colonies into approximately 2 ml of sterile saline solution. Inoculate the plates of tryptic soy agar with the bacterial suspension by a sterile swab, and streak a pure culture of S. aureus across each plate that had been inoculated. The culture media were then incubated for 18 to 24 hours in 5% CO2 at 37°C. Observe the suspected colonies of H. influenzae if the growth is seen in the agar plates as the colonies are close to the column of S. aureus and the growth is greater than that farther away from it [8].
2.3. DNA Extraction and Molecular Detection of H. influenzae
The DNA extraction method was performed according to the manufacturer’s protocols of the genomic DNA purification kit (Promega, USA). The pathogen suspected to be H. influenzae isolates was subjected to conventional PCR method to detect the specified gene amplification products of the omp6 gene for confirming isolation of H. influenzae, and the method was done as described previously [9]. The itemized sequences of the primer sets are illustrated in Table 1. A total volume of the reaction mixture (20 μl) contained 5 μl of DNA template, 3 μl of each primer, 5 μl of Green Master Mix (2×), and 4 μl of nuclease-free water. The thermal cycling condition usually starts the process with initial denaturation steps being carried out at 95°C for 10 minutes. Next, 30 cycles consisted of a denaturation step at 95°C for 30 seconds, annealing step at 55°C for 1 minute, and extension step at 72°C for 2 minutes. The final extension step was at 72°C for 5 minutes. The PCR amplification products were observed on a 1.5% agarose gel stained with ethidium bromide for 45 minutes at 70 Volt and visualized by UV transilluminator.
2.4. Molecular Detection of bexA and bexB genes in H. influenzae Isolates
The PCR technique was used for differentiating H. influenzae isolates into typeable and nontypeable using the primer pair with the amplicon size specific to bexA and bexB genes. The itemized sequences of the primer sets are illustrated in Table 1, as previously described in [10,11] with some modifications.
The thermal cycling condition usually starts the process with the “initial denaturation step being carried out at 95°C for 2 minutes. Next, 30 cycles consisted of a denaturation step at 95°C for 30 seconds, primer annealing step at 54°C for 30 seconds, and extension step at 72°C for 45 seconds. Then final extension step was at 72°C for 5 minutes.” The PCR amplification products were observed on a (1.5%) agarose gel stained with ethidium bromide for 45 minutes at 70 V and visualized by UV transilluminator.
3. RESULTS
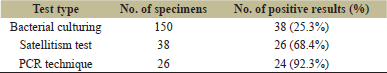
Among the 150 eye swabs obtained from patients suffering from conjunctivitis, it was found that only 38 isolates (25.3%) were approved as H. influenzae. Among these isolates, only 26 (68.4%) were positive by Satellitism test using discs of X and V factors which are required for growth of H. influenzae. To confirm isolation of the H. influenzae, these 26 isolates were subjected to further detection using a molecular PCR assay to identify of omp6 gene, and the results revealed that only 24 (92.3%) out of 26 isolates were positive for omp6 gene as shown in Figure 1.
The PCR method was used for differentiating all the 24 isolates of H. influenzae into both encapsulated (typeable) and nonencapsulated (nontypeable) H. influenzae using the primer pair with the amplicon size specific to bexA and bexB genes. The results found that all 24 isolates (100%) were negative (did not contain bexA and bexB) genes and are considered as nonencapsulated (nontypeable) isolates. The results of the PCR method for detection of the bexA gene are shown in Figure 2.
 | Table 1: The primer sequence and size of the product. [Click here to view] |
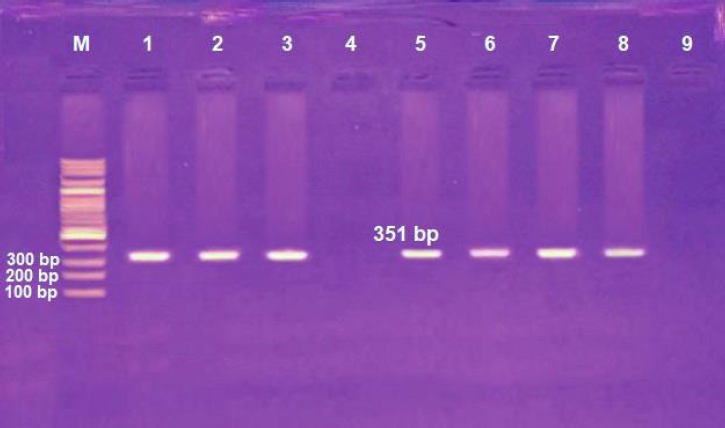 | Figure 1: Agarose gel electrophoresis of H. influenzae omp6 gene detection. Lane M: marker with 100 bp ladder and lanes 1–3 and 5–8: positive for omp6 gene, while lanes 4 and 9: negative for omp6 gene. Amplicon size: 351 bp. [Click here to view] |
 | Figure 2: Agarose gel electrophoresis of H. influenzae bexA gene detection. Lane M: marker with 100 bp ladder and lanes 1–9: negative for bexA gene. [Click here to view] |
 | Table 2: Profile the three diagnostic methods of H. influenzae isolates. [Click here to view] |
Also, the comparison and correlation of the three methods used in the diagnosis of H. influenzae are summarized in Table 2.
4. DISCUSSION
Haemophilus influenzae is the most common causative organism in children who suffer from acute bacterial conjunctivitis and mostly presents as conjunctivitis-otitis media syndrome coinciding with acute otitis media [12]. The most frequent cause of bacterial conjunctivitis is H. influenza. Complete or partial locus for capsule production is found in some strains of H. influenzae but lacks a functional “bexA” gene, which encodes for a protein involved in export of capsular polysaccharides to the cell surface. These strains are nontypeable H. influenzae (NTHI). Sinusitis, otitis media, conjunctivitis, and chronic obstructive pulmonary disease are mucosal infections, which are mainly caused by nontypeable of H. influenzae strains [13].
The findings of the present study showed differences in the rate of H. influenzae isolation, probably explained by a variety of factors such as the age of patients, sample size, season of sampling, the type of antibiotic used in treatment, which may be linked to H. influenzae expressing multiple virulence factors in numerous locations on the human body as its native host, and the type of detection methods that give sensitivity and specificity in more accuracy. In contrast, the results disagree with the previous results studies reported by Rai et al. [14], which showed that the cultural method might be used for H. influenzae isolates at a rate of around (18.05%) by using discs of X and V factors, while another study has revealed that the H. influenzae can be isolated at a rate of 27.16% by the use of selective media and discs of “X and V” factors [15]. Also, the previous results recorded in Iran found that the H. influenzae may be isolated from eye mucus of patients suffering conjunctivitis and other different sites at a prevalence rate (31.4%), depending on their morphology and growth requirement for “X and V” factors [16].
The molecular detection method was used to confirm the diagnosis of the H. influenzae isolates by detecting the presence of the omp6 gene. The omp6 gene was only detected in 24 isolates (92.3%) of H. influenzae obtained from patients with conjunctivitis. Despite the fact that H. influenzae has a number of OMPs such as P5 and P2, unlike P2, the P6 protein has a high homology at a rate of 97% in studies of type b and NTHI amino acids, indicating that it is strongly conserved and stable [17]. A previous study had proved that NTHi OMP P6 is a possible part of the vaccine for prevention of H. influenzae infections [18].
Additionally, the results revealed that all 24 isolates (100%) were noncapsulated (nontypeable) isolates by using the PCR method. The obtained result is higher than the result of the other research findings obtained from Ref. [19], which indicated that NTHI isolates account for 44%–68% of bacterial conjunctivitis cases in children. Other studies discovered that the existence or absence of the bexA gene determined if the isolate was encapsulated or nonencapsulated by molecular PCR assay; their findings showed that the rate of NTHi was about 93.5% [20].
5. CONCLUSIONS
NTHI was an important bacterium detected in eye swabs of patients suffering from conjunctivitis of both sexes. The use of the PCR method for the identification of H. influenzae isolates by detecting the omp6 gene was regarded as a faster, more sensitive, and more appropriate method than the use of traditional bacterial culturing methods and presumptive identification using X and V factor tests.
6. ACKNOWLEDGMENTS
The authors thank the reviewers for their evaluation of the manuscript.
7. AUTHORS’ CONTRIBUTION
All authors have made substantial contributions to the conception or design of the work, collection of the study samples, performing all the analysis and laboratory tests, interpretation of data for the work, preparation of the draft, revision of the article, and final approval of the version to be published.
8. CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
9. FUNDING
This work is self-funded.
10. ETHICAL APPROVALS
This study was approved by the Research Ethical Committee of both Ministry of Health and Environment and Ministry of Higher Education and Scientific Research in Iraq.
REFERENCES
1. Shekhawat NS, Shtein RM, Blachley TS, Stein JD. Antibiotic prescription fills for acute conjunctivitis among enrollees in a large United States managed care network. Ophthalmology 2017;124(8):1099–107. CrossRef
2. Oksola AO, Salako AO. Microbiological profile of bacterial conjunctivitis in Ibadan, Nigeria. Ann Ibadan Postgrad Med 2010;8(1):20–4. CrossRef
3. AL-Rubaey NKF, Sabri M, AL-Rubaey Q K. Isolation and characterization of bacteria from patients with conjunctivitis in Hilla Province. Med J Babylon 2007;4(1–2):36–44.
4. Power PM, Bentley SD, Parkhill J, Moxon ER, Hood DW. Investigations into genome diversity of Haemophilus influenzae using whole genome sequencing of clinical isolates and laboratory transformants. BMC Microbiol 2012;12:273. CrossRef
5. Osman KL, Jefferies JM, Woelk CH, Cleary DW, Clarke SC. The adhesions of non-typeable Haemophilus influenzae. Exp Rev Anti Infect Ther 2018;16(3):187–96. CrossRef
6. Willey JM, Sherwood LM, Woolverton CJ. Prescott, Harley and Klein’s microbiology. 7th edition, McGraw-Hill Higher Education, New York, NY, 2008.
7. MacFaddin, JF. Biochemical tests for the identification of medical bacteria. 3rd edition, The Williams and Wilkins-Baltimore, Baltimore, MD, 2000.
8. Xi Y, Wang H, Wang S, Wu X, Wang Z. Influencing factors in the satellitism test of Haemophilus influenzae and Haemophilus parainuenzae supplemented V factor by Staphylococcus aureus. Res Sq 2020;1–13. CrossRef
9. Ueyama T, Kurono Y, Shirabe K, Takeshita M, Mogi G. High incidence of Haemophilus influenzae in nasopharyngeal secretions and middle ear effusion as detected by PCR. J Clin Microbiol 1995;33(7):1835–8. CrossRef
10. Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of H. influenzae. J Clin Microbiol 1994;32(10):2382–6. CrossRef
11. Davis GS, Sandstedt SA, Patel M, Marrs CF, Gilsdorf JR. Use of bexB to detect capsule locus in H. influenza. J Clin Microbiol 2011;49(7):2594–601. CrossRef
12. Sugita G, Hotomi M, Sugita R, Kono M, Togawa A, Yamauchi K, et al. Genetic characteristics of Haemophilus influenzae and Streptococcus pneumoniae isolated from children with conjunctivitis-otitis media syndrome. J Infect Chemother 2014;20(8):493–7. CrossRef
13. Van Eldere J, Slack MPE, Ladhani S, Cripps AW. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis 2014;14(12):1281–92. CrossRef
14. Rai R, Kumar KU, Ramanath G. Haemophilus influenzae, an under diagnosed cause of respiratory tract infection. J Clin Diag Res 2012;6(3):385–7.
15. Das BK, Arora NK, Mathur P, Ostwal P, Mandal S, Kabra SK, et al. The nasopharyngeal carriage of Haemophilus influenzae. Indian J Paediatr 2002;69(9):75–7. CrossRef
16. Mojgani N, Rahbbar M, Taqizadeh M, Mohammadzadeh M. Biotyping, capsular typing and antibiotic resistance pattern of H. influenzae strain in Iran. Jpn J Infect Dis 2011;64(1):66–8.
17. Winn W, Allen S, Janda W, Koneman E, Procop G, Schreckenberger P, et al. Color atlas and textbook of diagnostic microbiology. 6th edition, Lippincott William and Wilkins, Philadelphia, PA, 2006.
18. Karalus RJ, Murphy TF. Purification and characterization of outer membrane protein P6, a vaccine antigen of non-typeable Haemophilus influenzae. FEMS Immunol Med Microbiol 1999;26(2):159–66. CrossRef
19. Block SL, Hedrick J, Tyler R, Smith A, Findlay R, Keegan E, et al. Increasing bacterial resistance in pediatric acute conjunctivitis (1997-1998). Antimicrobial agents and chemotherapy 2000;44(6):1650–4. CrossRef
20. Bonif?cio desilva MEN, Dasilva P, Medeiros MIC, Neme SN, Macedo C, Marin JM. Comparison of two slide agglutination serotyping and PCR-based capsule typing for the characterization of Haemophilus influenzae serotypes. Braz J Microbiol 2006;37:39–41. CrossRef