1. INTRODUCTION
Millets are the diverse group of cereals that produce small seeds. They include various annual food and fodder grasses such as foxtail millet (Setaria italica), pearl millet (Pennisetum glaucum), finger millet (Eleusine coracana), proso millet (Panicum miliaceum), barnyard millet (Echinochloa sp.), kodo millet (Paspalum scrobiculatum), etc. [1]. Among them, Foxtail millet (S. italica) belongs to tribe Paniceae under Poaceae family and the subfamily named Panicoideae [2]. It is generally named as Italian millet or German millet or Russian millet or Chinese millet or Hungarian millet; in India, it is commonly named as Kangni (Hindi) and is diploid (2n) in nature with 9 pairs of chromosomes [3,4]. The production cost is very inexpensive and also, they are pest-free crops. In India, foxtail millet is mainly cultivated in Andhra Pradesh, Karnataka, and Tamil Nadu. Foxtail millet has high nutritional value, in 100 g of foxtail millet grain contains protein 12.3 g, carbohydrates 60.9 g, vitamins 3.3 g, minerals 3.3 g, calcium 31 mg, and food energy nearly 323–350 K Cal [5]. Foxtail millets are rich in minerals such as iron, potassium, magnesium, zinc, and edible fiber (crude) and also free of gluten with a flat GI that helps reduce the production of glucose, result in lowering the risk of diabetes mellitus [1]. Also, these show strong resistance to several abiotic stresses, especially to drought and low nutrition cases. Diseases that affect foxtail millet include leaf and head blast disease [6]. Among biotic stressed, blast is a major disease which is caused by fungus called Magnaporthe grisea, affecting the leaf and head regions and reduces its yield. Fungi are one of the most effective pathogens that infect and lead to a vast crop yield losses annually [7].
Plants trigger defense response against invading pathogens upon the study of various immunogenic microbial signatures, such as microbe-associated molecular patterns, and PAMPs (pathogen-associated molecular patterns) [8]. The most significant component of pathogenesis-related proteins (PRs) functional families and are important to target genes to improve crop development is chitin [9]. Chitinases play a crucial role as a substrate for chitin with its hydrolase activity. These catalyze the hydrolytic cleavage of chitin’s β-1, 4 glycosidic bonds that are present in fungal cell walls [8]. During fungal infection, plants chitinases release chitin oligosaccharides which is a kind of PAMP, that activates the immune responses to protect plants from chitin-containing pathogens [10,11]. Studies reported that numerous chitinases are suggested to cleave arabinogalactan proteins and N-acetylglucosamine-containing glycoproteins in the plant cell walls, as a result, oligosaccharides are released that act as signal molecules triggering a defense response in plants [8].
Chitinases are differentiated mainly based on the catalytic domains it holds. They are divided into two families, GH18 and GH19 (Glycolyse Hydrolase) families [12], where GH18 and GH19 families further classified into five classes (class I–V). GH18 chitinases (class III and V) are broadly grouped in a variety of organisms such as plants, animals, fungi, and viruses [13], while GH19 chitinases (class I, II, IV) are located largely in higher plants [12], also responsible for major plants chitinolytic activity [14]. Besides pathogen infection, chitinases are also induced by various abiotic stresses like ethylene, salicylic acid, particles of pectin, heavy metals, wounds [15]. Effective studies are going on those chitinases that are expressed constitutively and specifically from different plant species expressed in organs such as flowers, leaves, seed, and roots, without any induction of pathogens or stress [16]. Thus, identification and studying these chitinases reveal their role and importance in Foxtail millet.
2. MATERIALS AND METHODS
2.1. Identification of Putative Chitinase Genes
Chitinase genes present in S. italica plant genome are retrieved by hidden markov model of Glyco_Hydro_18 (PF00704) and Glyco_Hydro_19 (PF00182) domains seed profiles are collected from Pfam 31.0 (https://en.wikipedia.org/wiki/Pfam, accessed date: 8th August 2020) [17]. These genes are collected using “PF00704” and “PF00182” as keywords (threshold E values ≤ 1.0) and also its amino acid length, gene sequence, coding region length are also obtained from the foxtail millet genomic database (Phytozome v12.1, https://phytozome.jgi.doe.gov/pz/portal.html#, accessed date: 28th August 2020). The entire sequences are checked for the presence of domains GH-18 and GH-19 using SMART (http://smart.embl-heidelberg.de/, accessed date: 1st September 2020). The molecular weights, isoelectric points, GRAVY values of these genes are predicted in ExPASy tool (https://web.expasy.org/protparam/, accessed date: 2nd September 2020) [18]. BaCello is used to predict the subcellular localization of all the genes [19], and the cleavage sites of signal peptide were predicted using SignalP [20].
2.2. Phylogenetic Analysis and Multiple Sequence Alignment
Evolutionary affiliation among the predicted chitinase genes was analyzed by aligning the full-length sequences of 65 chitinase proteins from S. italica (predicted sequences) and Arabidopsis thaliana (https://www.arabidopsis.org/) using Clustal W [21].
The unrooted neighbor-joining (NJ) phylogenetic tree was constructed based on Poisson correction using MEGA X [22]. Bootstrap analysis is employed using 1,000 replicates. The pair-wise gap deletion mode is done to make sure that the more divergent C-terminal domains can contribute to the topology of the NJ tree. Thus, all the positions that contain gaps and the missing data are deleted.
2.3. Visualization of Gene Structure and Chromosomal Localization
The coding sequence (CDS) and identical genomic sequence of all identified chitinase genes were utilized to predict the exon–intron structures using Gene Structure Display Server (GSDS v2.0) [23]. Chromosomal locations of chitinase genes were visualized by using MapGene v2.0 (http://mg2c.iask.in/mg2c_v2.0/) by using the gene annotation information accessible at the Phytozome database and chromosome data of the S. italica genome [24].
2.4. Identification of Conserved Domains, Motifs, and Active Site Analysis
MEGA X [22] and MEME suite 5.0.1 (Multiple Em for Motif Elicitation) [25] were used to identify conserved domains. Sequence alignment and editing are done by using BioEdit [26]. Multiple sequence alignment was done separately according to the homology of amino acid sequences in 4 groups in which group 1 comprised of chitinases of class I, class II, and class IV; group 2 contains numerous class I and IV chitinases; group 3 included class III chitinases which are in large number and group 4 includes only class V chitinase genes. MEME suite (http://meme-suite.org/index.html) is implied to identify the conserved motifs through which the classifications are confirmed.
2.5. Prediction of Cis-Acting Elements in Promoter Region
Cis-acting elements in the promoter region of chitinase genes were investigated using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) [27] by subjecting the 1,500 bp sequences upstream from the start codon which is retrieved from the Phytozome database [17].
2.6. Gene Ontology Annotation of Chitinase Genes
To obtain the Ontology of Chitinase Genes, the Blast2GO tool [31]was used where the sequences are imported onto the Blast2GO tool’s workspace and screened using BLASTP. Then the gene mapping and annotation are studied by using InterproScan, whereas the biological processes (BP), cellular components (CC), and metabolic pathways were predicted by using identified GO terms and represented in statistical chart form.
2.7. 3-D Structural Analysis of Chitinase Genes
The 3-dimentional structure of the chitinase proteins are analyzed and constructed using Phyre V 2.0 server [28]. There should be a 75% and more alignment coverage and 100% thresholds confidence so that the quality of the model structure is maintained. MEMSAT-SVM prediction method is used to predict the transmembrane helix and membrane topology of all the chitinases.
3. RESULT
3.1. Identification of Chitinase Genes
A total of 46 putative chitinase genes are identified (6 sequences considered as putative chitinases-like proteins— (CLP) as a result of the search in the phytozome database among the Foxtail millet genome. The length of these genes ranges from 222 to 1,038 residues, the protein molecular weights range between 23.98 and 52.24 kDa, and isoelectric points (pI) vary from 4.28 to 9.92. Among those 40 chitinases, 31 are predicted to be located in secretory pathway (SP), 3 are localizing in the chloroplast (C) and 6 in the nucleus (N) (Table 1). Chitinases are classified into 5 classes within 2 families (GH_19 and GH_18 families) [12] where GH-19 includes Class-I, II, and IV chitinases, GH_18 includes Class-III and V chitinases. Class-I has highly conserved N-terminal cysteine-rich region that involves in chitin-binding, and seen in two sequences, i.e., SICHI010, SICHI011. Class II lacks both the N-terminal cysteine-rich region and the C-terminal extension, which have a catalytic domain that is observed among 10 sequences [29]. Class IV chitinases resemble the class I chitinases with a very similar main structure but are significantly smaller due to four deletions distributed along with the chitin-binding domain and the catalytic region which are satisfied by 7 sequences, i.e., SICHI002, SICHI004, SICHI007, SICHI014, SICHI015, SICHI016, SICHI018. While in the GH_18 family, 19 sequences belong to Class-III and Class-V into two based on c-terminal extension and catalytic domain position [15].
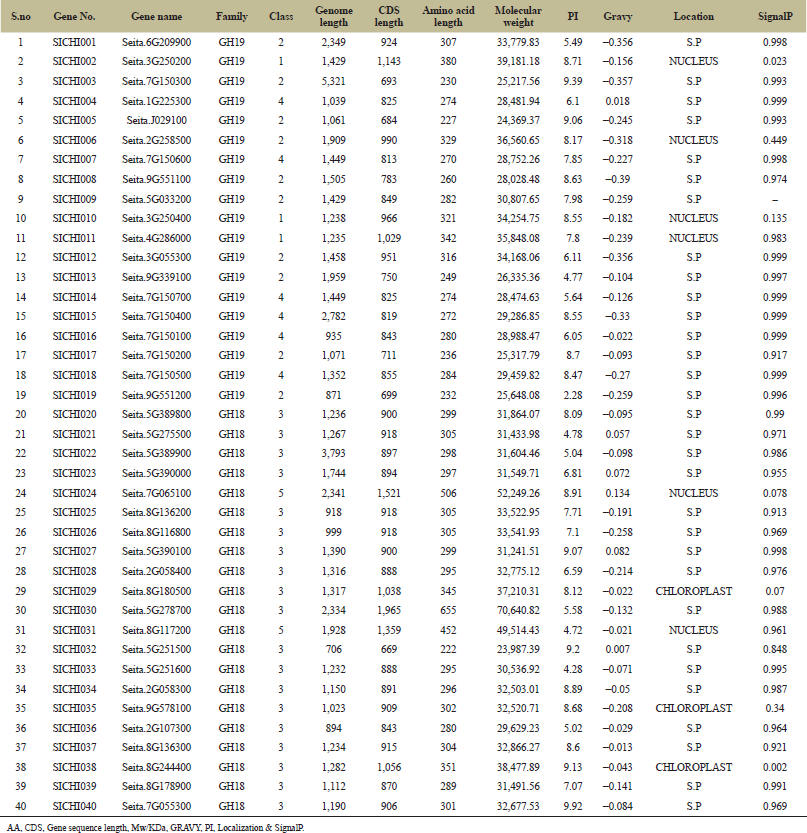 | Table 1: Physicochemical properties of predicted chitinase genes in Foxtail millet.z [Click here to view] |
3.2. Phylogenetic Analysis and Multiple Alignment
To evaluate the evolutionary relationship of the chitinase genes in the foxtail millet, A. thaliana chitinase sequences are used as reference sequences to generate the phylogenetic analyses based on the NJ method using MEGA X. Total 64 chitinase proteins are predicted in both A. thaliana and S. italica and categorized into two important evolutionary branches which determine the GH19 and GH18 families, which were furthermore divided into five groups, class-I–V. GH18 family comprises the classes III and V with a total of 31 sequences, while GH19 family comprises the classes I, II, and IV with 33 sequences altogether. Class III is the largest class with 20 sequences representing nearly 31% of the total number of chitinase genes, while class I is the smallest, with only 3 sequences that represent only 4% approximately. The remaining classes-II, IV, and V share 21.87%, 25%, and 17.18% of chitinases, respectively. When it comes to Foxtail millet, class III shares a greater number of sequences, i.e., 19, and both the classes I and V share only 2 sequences in each. The phylogenetic tree represented below shows that the GH18 and GH19 domains are separated into two different clades (Fig. 1).
3.3. Visualization of Chitinase Genes on Chromosomes
Out of all the 40 chitinases, 12 chitinases have zero introns, 6 chitinases with 2, and the remaining 18 with only one intron, including SICHI015 with a large intron (Fig. 2). Chromosomal positions are determined by the presence of the maximum numbers of genes, i.e., nine are spotted on the 7th chromosome followed by eight on the 5th chromosome and seven genes on the 8th chromosome. Four each on 2nd and 9th chromosomes, three genes are identified on 3rd, two on 1st, and one gene each on 4th and 6th chromosomes (Fig. 3). Some clusters are also seen like on 5 and 7 chromosomes with two clusters each. Two genes (SICHI022 and SICHI0027) and (SICHI020 and SICHI0023) form two clusters on chromosomes 5, where all the genes belong to class-III. Two genes from class IV (SICHI0007, SICHI0015), one from class II (SICHI0017) form to a single cluster and one more cluster is formed by four genes in which three genes (SICHI0014, SICHI0016, SICHI0018) belong to class-IV and one from class-II (SICHI0003) on chromosomes 7.
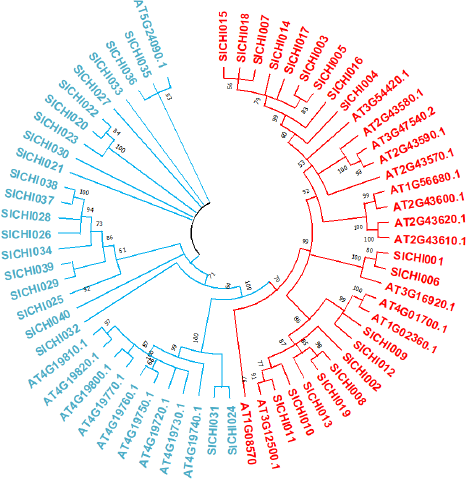 | Figure 1: Phylogenetic tree built using the NJ method in MEGA X. Boot strap values are from 1,000 replications. GH19 family sequences got separated on right side clade, GH18 family sequences on left. The value at the nodes represents bootstrap percentage. [Click here to view] |
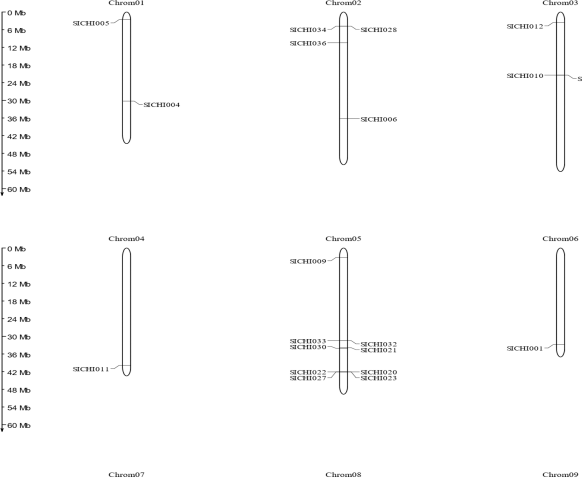 | Figure 2: All the information about the chromosome number and size are identified and represented in the above figure. [Click here to view] |
3.4. Conserved Domains and Active Site Analysis
Foxtail millet chitinases contain the conserved domain structures according to the previously identified chitinases in plant [12]. Studying the gene sequences shows the existence of the catalytic domain GH19 (PF00182) with one or two active sites that were represented as CHITINASE_19_1 (PS00773) and/or CHITINASE_19_2 (PS00774) (Fig. 4a) and I and IV classes of chitinases possess a domain that binds chitin that is CHIT_BIND_I_1 (PS00026) located at N-terminal (Fig. 4b). Another catalytic domain GH-18 (PF00704), i.e., CHITINASE_18 (PS01095) with classes III and V active site (Fig. 4c). Functional site analysis of these sequences was done using ExPASyprosite [18] results that out of 46, in 6 sequences catalytic domains are devoid of active sites responsible for their chitinolytic activity also had incorrect amino acid sequences at their active sites. Hence, they are classified as putative CLPs. Multiple sequence alignment of foxtail millet chitinases is done in four groups. Sequence alignment is done using ClustalW and edited using BioEdit. The diversification of chitinase proteins and its conserved motifs are examined using MEME suite, where 30 conserved motifs are found in the chitinase proteins (Fig. 4d). Such similar motif compositions of each group provide added proof in supporting the phylogenetic analyses results but also presumed functional significance.
3.5. Cis-Acting Elements Prediction in Promoter Region
The transcriptional regulation of chitinase genes was done by several phytohormones and pathogens [30]. To interpret the probable regulation mechanisms of SICHI genes, the cis-acting elements situated in the promoter sequence are predicted using Plant-CARE [27]. Many elements were identified out of which 18 elements were more common in all the genes, which functions in several hormone regulations and stress-related elements (Table 2). In our results, nearly 19 elements that are related to response to stress [TATA box, CAT, CAAT, G-box, W-box, ARE, GARE motif, GT I-motif, TC-rich repeat, MYB binding site (MBS), CGTCA-motif, SKn-I, ACI, WUN-motif, dehydartion responsive element, HD-ZIP, low temperature induced (LTR), MBS, stress responsive elements (STRE)]) and few hormones like TGACG-motif, ethylene-responsive element, ABA-responsiveness (ABRE), Auxin-responsiveness, and SA-responsiveness elements were observed
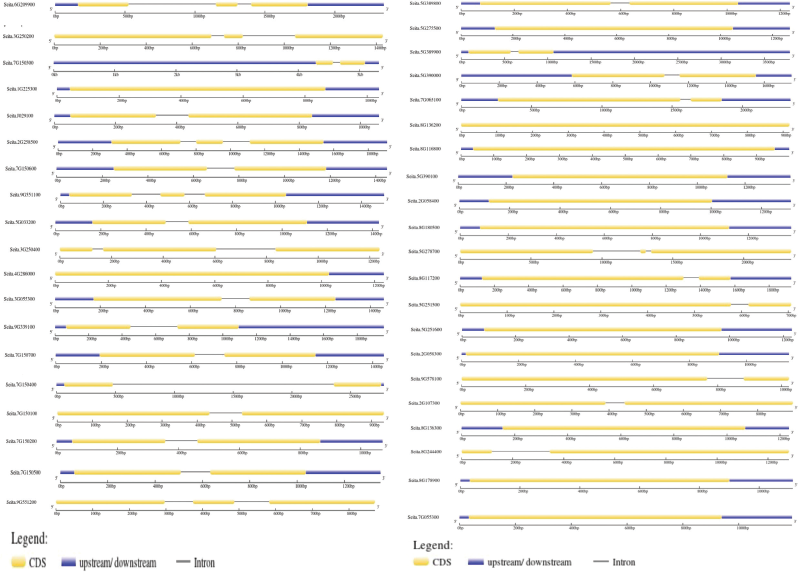 | Figure 3: (a, b) The intron–exon structure of all the 40 chitinase genes in Foxtail millet. The CDS is indicated by a yellow horizontal bar, the untranslated regions are indicated by a blue horizontal bar, and introns are determined by the black line. (a) determines GH19, (b) determines GH18 domain family genes. [Click here to view] |
3.6. Gene Ontology Annotation of chitinases Genes
Gene Ontology of 64 chitinases genes results that these are associated in various bpBP, molecular functions (MF), ccCC [31] and are represented in Figure 5. Among various BPb, different metabolic processes and responses to stimuli (under stress conditions) are mostly associated. Hydrolase activities and binding activities are mainly observed in MF. Most of the CC are found in extracellular region and the remaining are distributed in membrane regions.
3.7. 3-D Structural Analysis of Sichi Genes
The 3-dimensional structure of chitinase genes helps us by understanding insights the chitin-binding domain by providing information that is usually hard to get experimentally. The 3-D supermolecule structures were sculptured for all the I–V chitinases using the Phyre V 2.0 server [28] for understanding the structural properties of foxtail millet chitinase genes. Protein models were built under default parameters [32]. Moreover, these predicted 3D protein structures are considered highly stable and give a basic understanding of the alpha chains and beta sheets in percentages form. Classes-I, IV, and V chitinases were able to predict membrane topology, which provides the signal peptide with a varied length of amino acids, at N-terminal that supports delivering the protein at its target site, and gets cleaved-off once it reaches its destination. All these chitinases are cytoplasmic and their action is extra-cellular (Fig. 6).
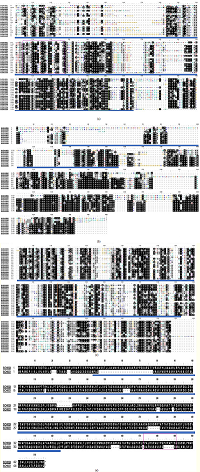 | Figure 4: (a) Class-I, II, & IV of GH-19 family gaps were deleted, the chitinase 19_1 signature PS00773 [Cx[4,5]-FY-(ST)-x(3)-(FY)-(LIVMF)-xAx(3)-(YF)-x(2)-F—(GSA)] is indicated by violet box (205–227 aa) and Chitinase 19_2 signature PS00774 [(LIVM)-(GSA)-Fx-(STAG)(2)-(LIVMFY)-W-(FY)-W-(LIVM)] is indicated by green box(343–357aa). Yellow line indicates the active site of GH-19 (75–365 aa). (b) Class-I and IV, the chitin-binding domain (26–155 aa) is indicated by red line, as this is only present in class I & IV. (c) Class-III and V from the GH-18 family gaps were deleted, the Chitinase_18 signature PS01095 [(LIVMFY)-(DN)-G-(LIVMF)-(DN)-(LIVMF)-(DN)-x-E ] is indicated by orange box (224–232 aa) and the GH-18 active site is underlined by blue line (28–292 aa). Gaps were deleted. (d) Class-V from GH-19 family, the pink box (473–490) determines CRYSTALLYN_BETAGAMMA signature PS00225 [(LIVMFYWA)-{DEHRKSTP}-(FY)-(DEQHKY)-x(3)-(FY)-x-G-x(4)-(LIVMFC-ST)] and the GH-18 active site is underlined by blue line. [Click here to view] |
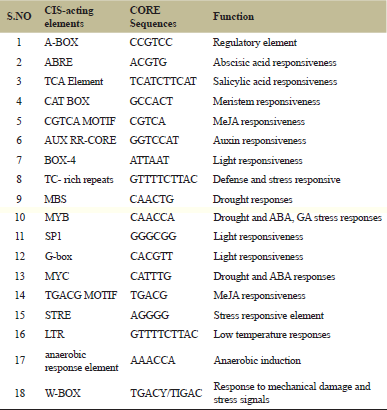 | Table 2: Predicted CIS-acting elements which are more commonly identified along with their functions and core sequences. [Click here to view] |
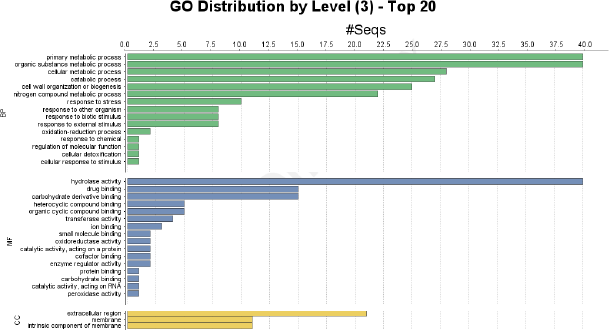 | Figure 5: Chitinase gene ontology analysis and the GO terms which are involved in BP, MF, & CC. [Click here to view] |
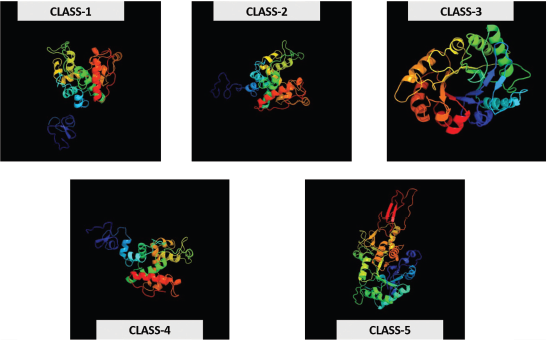 | Figure 6: Classes I–V predicted 3-D protein structures of foxtail millet chitinases. [Click here to view] |
4. DISCUSSION
Foxtail millet is a versatile and stable food crop globally and is cultivated widely in arid and semi-arid areas of China, India, Japan, and some part of South and North America [33]. Among all the millets, Foxtail millet is the second most cultivated and next to Pearl millet (FAOSTAT 2005; http://faostat.fao/org/) it has a wide tolerance to abiotic stress. However, plants have a continuous threat from pathogen infections and affect their growth and yield. During such stress conditions, plants activate their defense system and start to produce PR proteins that act as antimicrobial compounds [34]. Among various PR proteins, chitinases play a major role and exhibit anti-fungal activity by catalyzing the hydrolysis of chitin which is the central element of the fungal cell wall [18]. Research on chitinase genes is mainly done in some of the major crops like A. thaliana, Oryza sativa, and Zea mays. In the present investigation, chitinase genes identification studies are done in S. italica which is most prone nature towards blast disease.
Among 46 chitinase genes identified, 6 were expelled as they lack catalytic amino acids and missing motifs. Thus, 40 chitinase genes were finalized in the foxtail millet genome and the number of chitinases is higher than that of A. thaliana, Brassica rapa [35], pigeon pea, and Ammopiptanthus nanus, however significantly lesser than Solanum lycopersicum [36]. Further, these 46 genes were differentiated in to two groups and five different classes (I–V) in which 21 genes are grouped under GH-18 family domain and the remaining 19 under GH-19 family. All these genes were distributed on nine chromosomes. Among them most of the chitinases were located on chromosome 5, 7, and 8, and only one chitinase was observed on chromosomes 1, 4, and 6 each and some clusters were present on chromosomes 5 and 7. In A. nanus, all the genes were distributed on 9 chromosomes in which clusters were observed on all the chromosomes expect on chromosome 7 [37], whereas in S. lycopersicum, the chitinase genes were sited on 12 chromosomes in which a total of 34 genes were found to be clustered onto the chromosomes 1, 2, 3, 7, 10, and 11 [36].
Chitinases of Class III and V were having an additional Lysozymal activity and showed grater similar to fungal, yeast, and bacterial chitinases [9]. Some theoretical studies reported that the class III chitinases have a different and bigger role than compared to other classes. The present investigation on S. italica genome also shows that class III was the largest class with 19 chitinases and similar to that of A. nanus, Cucumis sativus was also significantly higher than A. thaliana [37]. In Pteris ryukyuensis two N-terminal LysM domains on class-III chitinases contribute to have an antifungal activity where the LysM helps in binding to chitin in the cell wall of fungi [38]. They observed an abnormal expression of class-III chitinases genes in sugarcane, enhancing its biotic and abiotic stress tolerance [39]. Besides its theoretical import, more research has to be done to know its biological significance and difference between class-III and other plant chitinases.
Studies reported that the density of introns in chitinase genes was inversely proportional to stress responses regulation. Stress-related genes possess very less number of introns so that they regulate rapidly during stress conditions [40]. The number of introns in chitinase genes of S. italica is very low. These chitinase gens generally maintain particular amino acids for the catalytic activity, where in GH-18 family genes there was a major reduction of chitinase activity when they got mutated with less Glu(E)-Trp(W) and Asp(D)-Trp(W) residues along with more repeats of Met(M), Leu(L), Gly(G), Val(V), and Ile(I) [36]. In GH-19 family genes, the presence of two consecutive Glu (E) residues and some repeats of Asp(D), Thr(T), or Ser(S) in the catalytic region [41]. In A. nanus, out of 28 chitinase genes, 25 of these genes had two or fewer introns [42] and in B. rapa genome 32 chitinase genes out of 33 were identified to have two or fewer introns [35].
Chitinases were involved in many tolerant/adaptive responsive activities in stress conditions that regulated transcriptionally with the help of many Cis-regulatory elements analyzed using the PlantCARE database. Elements like MBS, MYB, MYC, and LTR were more commonly observed in promoter site of most of the chitinases genes in S. italica that were involved in response to low temperature and drought stress conditions. MYC, LTRE-1, and MYB were also observed in A. nanus [37]. One of the most frequently occurring cis-acting elements in promoter regions of foxtail millet is the TGACG motif and TATA-box, which were present in promoters of most of the chitinase genes. The C-repeat binding factors operate an important role in plants to tolerate cold responses and adaptive to the new environment [43]. Also, TC-rich repeats have a predominant role in developing strong defensive responses and W-box interferes in the fungal elicitor-induced gene expression by associating with WRKY1 in the parsley family [44]. Several studies identified that ethylene (ERE-ethylene-responsive) was a key hormone in the induction of chitinase genes [45]. Other hormones like ABRE, TCA, and TGACG-motif were predicted to regulate the chitinase genes transcription [46] and some stress-responsive elements such as STRE, W-box, and WUN-motif mediate pathogen or elicitor inducible transcription of chitinases [47,48].
Gene ontology results have shown different GO terms out of which primary metabolic process, cellular metabolic process was high and expressed in all the genes along with, response to stress and other organisms also share a moderate level of expression in most of them, which might be useful in defensive response to fungal pathogens. In MF, genes showed the elevated expression of hydrolase activity and an average expression of catalytic activity in all the genes.
The protein structures of randomly selected genes, each from all the five classes, made a clear idea about the alpha, beta, and transmembrane helices percentage that helps determine the strength of the hydrogen bonds and the protein structure. The knowledge of transmembrane (TM) helices can be valuable in limiting the possibilities of tertiary structure formation for the given protein as well as in predicting the function [49]. As the TM helices for both Class-II and III were missing and made as a limitation for predicting the function of these chitinases whether they help in inducing the defense response against the fungal pathogens.
5. CONCLUSION
Total forty chitinase family genes were identified from the foxtail millet genome, in which, 21 were group into the GH-18 subfamily, and GH-19 under GH-19 subfamily. Class-I has 2, Class-II with 10, Class-III has 19, Class-IV has 7, and finally, Class-V has 2 chitinases genes in S. italica. A greater number of Class-III genes were observed comparatively, inferring that Class-III genes with fewer non-coding regions may be magnified in signifying their involvement in stress response. Gene ontology expression study of chitinases under biological process, MF, and CC GO terms, the prediction of the cis-acting regulatory elements in the promoter region of the genes, helps discover chitinases with key roles in the abiotic stress responses and defense against pathogens in S. italica. 3-D structure prediction was also done for five random genes from every class along with alpha, beta, and transmembrane helices. The research and results could provide solid data for assuming the functions of chitinases in foxtail millet and might help classify the chitinase genes, which can be used for both pathogen control and improvement of abiotic stress tolerance in crop improvement.
6. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the international committee of medical journal editors (ICMJE) requirements/guidelines.
7. FUNDING
There is no funding to report.
8. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
9. ETHICAL APPROVALS
Not applicable.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
11. REFERENCES
1. Lata C, Gupta S, Prasad M. Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit Rev Biotechnol 2013;33(3):328–43. CrossRef
2. Prasad M. The foxtail millet genome. Springer, Heidelberg, Germany, 2017. CrossRef
3. Lata C, Gupta S, Prasad M. Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses. Crit Rev Biotechnol 2013;33(3):328–43. CrossRef
4. Panaud O. Foxtail millet. In: Chittaranjan K (ed.). Cereals and millets. Springer, Heidelberg, Germany, pp 325–32, 2006. CrossRef
5. Vanithasri J, Kanchana S, Hemalatha G, Vanniarajan C, Sahulhameed M. Role of millets and its importance in new mellinium. Int J Food Sci Technol 2012;2(1):35–47.
6. Pennisi E. Armed and dangerous. Science 2010;327(5967):804–5. CrossRef
7. Shuping DSS, Eloff JN. The use of plants to protect plants and food against fungal pathogens : a review. Afr J Tradit Complement Altern Med 2017;14(4):120–7. CrossRef
8. Newman MA, Sundelin T, Nielsen JT, Erbs G. MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 2013;4:139. CrossRef
9. Grover A. Plant chitinases: genetic diversity and physiological roles. Crit Rev Plant Sci 2012;31(1):57–73. CrossRef
10. Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci 2007;104(49):19613–8. CrossRef
11. Legrand M, Kauffmann S, Geoffroy P, Fritig B. Biological function of pathogenesis-related proteins: four tobacco pathogenesis-related proteins are chitinases. Proc Natl Acad Sci 1987;84(19):6750–4. CrossRef
12. Passarinho PA, de Vries SC. Arabidopsis chitinases: a genomic survey. The Arabidopsis book/American Society of Plant Biologists, Rockville, MD, 2002. CrossRef
13. Arakane Y, Muthukrishnan S. Insect chitinase and chitinase-like proteins. Cell Mol Life Sci 2010;67(2):201–16. CrossRef
14. Tyler L, Bragg JN, Wu J, Yang X, Tuskan GA, Vogel JP. Annotation and comparative analysis of the glycoside hydrolase genes in Brachypodium distachyon. BMC genomics 2010;11(1):1–21. CrossRef
15. Graham LS, Sticklen MB. Plant chitinases. Can J Bot 1994;72(8):1057–83. CrossRef
16. Zhong R, Kays SJ, Schroeder BP, Ye ZH. Mutation of a chitinase-like gene causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 2002;14(1):165–79. CrossRef
17. Yang R, Chen M, Sun JC, Yu Y, Min DH, Chen J, et al. Genome-wide analysis of LIM family genes in Foxtail Millet (Setaria italica L.) and characterization of the role of SiWLIM2b in drought tolerance. Int J Mol Sci 2019;20(6):1303. CrossRef
18. Bartholomew ES, Black K, Feng Z, Liu W, Shan N, Zhang X, et al. Comprehensive analysis of the chitinase gene family in Cucumber (Cucumis sativus L.): from gene identification and evolution to expression in response to Fusarium oxysporum. Int J Mol Sci 2019;20(21):5309. CrossRef
19. Pierleoni A, Martelli PL, Fariselli P, Casadio R. BaCelLo: a balanced subcellular localization predictor. Bioinformatics 2006;22(14):e408-16. CrossRef
20. Nielsen H. Predicting secretory proteins with SignalP. In Kihara D (ed.). Protein function prediction. Humana Press, New York, NY, pp 59–73, 2017. CrossRef
21. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994;22(22):4673–80. CrossRef
22. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 2018;35(6):1547–9. CrossRef
23. Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 2015;31(8):1296–7. CrossRef
24. Ni X, Xia Q, Zhang H, Cheng S, Li H, Fan G, et al. Updated foxtail millet genome assembly and gene mapping of nine key agronomic traits by resequencing a RIL population. GigaScience 2017;6(2):giw005. CrossRef
25. Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 2006;34(Suppl_2):W369–73. CrossRef
26. Hall T, Biosciences I, Carlsbad C. BioEdit: an important software for molecular biology. GERF Bull Biosci 2011;2(1):60–1.
27. Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 2002;30(1):325–7. CrossRef
28. Kelley LA, Sternberg MJ. Protein structure prediction on the web: a case study using the Phyre server. Nat Protoc 2009;4(3):363. CrossRef
29. Iseli B, Boller T, Neuhaus JM. The N-terminal cysteine-rich domain of tobacco class I chitinase is essential for chitin binding but not for catalytic or antifungal activity. Plant Physiol 1993;103(1):221–6. CrossRef
30. Grover A. Plant chitinases: genetic diversity and physiological roles. Crit Rev Plant Sci 2012;31(1):57–73. CrossRef
31. Lin S, Qiao N, Chen H, Tang Z, Han Q, Mehmood K, et al. Integration of transcriptomic and metabolomic data reveals metabolic pathway alteration in mouse spermatogonia with the effect of copper exposure. Chemosphere 2020;256:126974. CrossRef
32. Mir ZA, Ali S, Shivaraj SM, Bhat JA, Singh A, Yadav P, et al. Genome-wide identification and characterization of Chitinase gene family in Brassica juncea and Camelina sativa in response to alternaria brassicae. Genomics 2020;112(1):749–63. CrossRef
33. Prasad M. The foxtail millet genome. Springer International Publishing, Cham, Switzerland, 2017. CrossRef
34. Hammerschmidt R. Phytoalexins: what have we learned after 60 years? Annu Rev Phytopathol 1999;37(1):285–306. CrossRef
35. Chen J, Piao Y, Liu Y, Li X, Piao Z. Genome-wide identification and expression analysis of chitinase gene family in Brassica rapa reveals its role in clubroot resistance. Plant Sci 2018;270:257–67. CrossRef
36. Cao J, Tan X. Comprehensive analysis of the chitinase family genes in tomato (Solanum lycopersicum). Plants 2019;8(3):52. CrossRef
37. Cao S, Wang Y, Li Z, Shi W, Gao F, Zhou Y, et al. Genome-wide identification and expression analyses of the chitinases under cold and osmotic stress in Ammopiptanthus nanus. Genes 2019;10(6):472. CrossRef
38. Onaga S, Taira T. A new type of plant chitinase containing LysM domains from a fern (Pteris ryukyuensis): roles of LysM domains in chitin binding and antifungal activity. Glycobiology 2008;18(5):414–23. CrossRef
39. Su Y, Xu L, Fu Z, Yang Y, Guo J, Wang S, et al. ScChi, encoding an acidic class III chitinase of sugarcane, confers positive responses to biotic and abiotic stresses in sugarcane. Int J Mol Sci 2014;15(2):2738–60. CrossRef
40. Jeffares DC, Penkett CJ, Bähler J. Rapidly regulated genes are intron poor. Trends Genet 2008;24(8):375–8. CrossRef
41. Kawase T, Saito A, Sato T, Kanai R, Fujii T, Nikaidou N, et al. Distribution and phylogenetic analysis of family 19 chitinases in actinobacteria. Appl Environ Microbiol 2004;70(2):1135–44. CrossRef
42. Bartholomew ES, Black K, Feng Z, Liu W, Shan N, Zhang X, et al. Comprehensive analysis of the chitinase gene family in Cucumber (Cucumis sativus L.): from gene identification and evolution to expression in response to Fusarium oxysporum. Int J Mol Sci 2019;20(21):5309. CrossRef
43. Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci 1997;94(3):1035–40. CrossRef
44. Eulgem T, Rushton PJ, Schmelzer E, Hahlbrock K, Somssich IE. Early nuclear events in plant defence signalling: rapid gene activation by WRKY transcription factors. EMBO J 1999;18(17):4689–99. CrossRef
45. Taira T, Toma N, Ichi M, Takeuchi M, Ishihara M. Tissue distribution, synthesis stage, and ethylene induction of pineapple (Ananas comosus) chitinases. Biosci Biotechnol Biochem 2005;69(4):852–4. CrossRef
46. Yamamoto S, Nakano T, Suzuki K, Shinshi H. Elicitor-induced activation of transcription via W box-related cis-acting elements from a basic chitinase gene by WRKY transcription factors in tobacco. Biochim Biophys Acta Gene Struct Expr 2004;1679(3):279–87. CrossRef
47. de las Mercedes Dana M, Pintor-Toro JA, Cubero B. Transgenic tobacco plants overexpressing chitinases of fungal origin show enhanced resistance to biotic and abiotic stress agents. Plant Physiol 2006;142(2):722–30. CrossRef
48. Gao Y, Zan XL, Wu XF, Yao L, Chen YL, Jia SW, et al. Identification of fungus-responsive cis-acting element in the promoter of Brassica juncea chitinase gene, BjCHI1. Plant Sci 2014;215:190–8. CrossRef
49. Ganapathiraju M, Balakrishnan N, Reddy R, Klein-Seetharaman J. Transmembrane helix prediction using amino acid property features and latent semantic analysis. BMC Bioinformatics 2008;9:S1–4. CrossRef