The increasing emergence of methicillin-resistant staphylococci (i.e., methicillin-resistant Staphylococcus aureus [MRSA] and methicillin-resistant coagulase-negative staphylococci [MRCoNS]) has become a threat globally for both human and animal populace. Phenotypic detection of MRSA and MRCoNS is a less sensitive and time-consuming approach which affects the treatment outcome. Thus, a rapid and accurate method is needed for an early diagnosis of MRSA/MRCoNS infections. The present study aimed at standardization and validation of a multiplex polymerase chain reaction (mPCR) assay to detect genus Staphylococcus (16s rRNA gene) and methicillin-resistance determinants (mecA and mecC genes) simultaneously. The assay characteristics were evaluated against 53 well characterized strains comprising of 40 Staphylococcus and 13 non-Staphylococcus strains. Among Staphylococcus strains, 32 were mecA positive and one strain was mecC positive. The lower limit of detection of the mPCR assay was 1ng/mL (Genome copies: 16S rRNA = 1.1 × 109; mecA = 3.17 × 109; mecC = 1.6 × 109), with analytical sensitivity and specificity of 100%. The mPCR assay developed in the study is useful for rapid and accurate diagnosis of MRSA/MRCoNS infections. The assay can be an important diagnostic as well as surveillance tool to investigate the emergence and dissemination of methicillin-resistant staphylococci which is of both clinical and public health significance.
Venugopal N, Ganaie F, Mitra S, Tewari R, Dey TK, Ojha R, Shome R, Shome BR. Development and validation of multiplex polymerase chain reaction for concomitant detection of genus Staphylococcus and clinically relevant methicillin resistance determinants. J App Biol Biotech. 2020;8(05):1-6. DOI: http://dx.doi.org/10.7324/JABB.2020.80501
1. INTRODUCTION
Members of genus Staphylococcus are widespread globally colonizing 20–30% of the human population [1]. They are known to cause wide array of infections including mild skin infection, sepsis, and even life-threatening bacteremia in certain cases [2]. Genus Staphylococcus comprises of two groups, namely, coagulase-positive Staphylococcus and the emerging coagulase-negative staphylococci (CoNS) [3]. Apart from human medicine, Staphylococcus spp. has also gained importance in veterinary medicine affecting livestock and poultry. The transmission of drug resistant strains from animals to humans and vice-versa can have a potential impact on the public health. There are evidences of clonal transmission of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-resistant CoNS (MRCoNS) between humans and cows, and also these strains may spread clonally within a herd of dairy cows [4,5].
In Staphylococcus, methicillin resistance is primarily as a result of altered penicillin binding protein (PBP2a) that shows very less affinity for all beta-lactam antibiotics. PBP2a is regulated by mecA gene. Identification of mecA gene among staphylococcal isolates is considered as the gold standard for the presence of methicillin resistance [6,7]. The chromosomal mediated mecA gene is harbored on a large mobile genetic element called SCCmec which are the known vectors that transfer the resistant genes among various Staphylococcus species. At present, international working group on classification of staphylococcal cassette chromosome has recognized 11 different types of SCCmec elements [8]. The existence of closely related SCCmec elements in S. aureus and CoNS implies horizontal transfer of SCCmec [9].
In 2011, a study from England on bovine mastitis reported a novel mecA homolog in MRSA isolates carrying SCCmec type XI. The homologous gene was initially called mecALGA251, but later renamed as mecC [10]. This novel mecC showed approximately 69% identity to mecA at the DNA level and can colonize, causing diseases in humans and other host species [11]. In contrast to these chromosomally mediated mecA and mecC genes for methicillin resistance, mecB gene originally described as mecAm, was identified in transposon mec complex (Tn6045) of Macrococcus caseolyticus [12]. However, in Macrococcus canis, SCCmec carrying mecB gene independent of Tn6045 was identified [13,14]. In addition, mecD gene, which share above 66% of nucleotide identity with mecB gene, was recently found in M. caseolyticus isolates of bovine and canine origin [15].
Although, there are numerous conventional phenotypic approaches for unmasking the methicillin resistance among staphylococci [16,17] like-oxacillin agar screen method, determination of minimum inhibitory concentration using agar dilutions, broth dilutions and E tests, cefoxitin, and oxacillin disk diffusion tests; however, discrepancies in the accuracy of these methods make them unreliable. Apart from being less sensitive and time consuming, the phenotypic expression of drug resistance depends on various conditions, i.e., growth media, temperature, and osmolarity. These variables further add to the limitation of these methods [18,19]. Conversely, molecular approach offers a rapid, accurate strategy with increased sensitivity and specificity for the identification of these drug resistant pathogens. Herein, we developed a novel multiplex polymerase chain reaction (mPCR) for the concurrent detection of Staphylococcus genus, and the methicillin resistance determinants mecA and mecC. The timely detection of methicillin resistance among staphylococci will be valuable for the quick diagnosis of the ailment caused by drug-resistant strains.
The selection of strains was done in such a way so that our assay is tested against multiple positive and negative controls for the genus Staphylococcus and methicillin resistance. The panel included 40 well characterized strains which served positive control for genus staphylococci. Out of these strains, 32 were used as a positive control for mecA gene and one S. saprophyticus strain (BH32) identified in our laboratory previously was used as a positive control for mecC gene. (GenBank Accession No: MH448900). The mecA positive staphylococcal strains were isolated from cattle milk (n = 23), cattle nasal (n = 1), cattle wound (n = 1), extramammary site (n = 2), and hand swab of animal handlers (n = 3). The additional two mecA positive strains were acquired from ATCC (American type culture collection), namely, ATCC 33591 (MRSA), and ATCC 43300 (MRSA). The mecC positive strain was isolated from buffalo milk.
In addition, we tested our assay with a negative control panel (n = 13), which covered different genera of Gram-positive and Gram-negative bacteria, namely, Streptococcus agalactiace, Streptococcus uberis, Streptococcus dysgalactiae, Lactobacillus acidophilus, Lactococcus lactis, Leuconostoc mesenteroides, Micrococcus aurantiacus, Enterococcus faecalis, Aerococcus viridans, Streptococcus bovis, Escherichia coli, Shigella sonnei, and Klebsiella pneumoniae. This panel was used a negative control for all the targets included in the mPCR assay. The other ATCC strains involved in the standardization of mPCR assay are detailed in Table 1.
Table 1: Evaluation of multiplex PCR against nine staphylococcal and 13 non-staphylococcal ATCC strains
| Strain name | Species | 16S rRNA | mecA | mecC |
|---|---|---|---|---|
| ATCC 25923 | Staphylococcus aureus | + | − | − |
| ATCC 25904 | Staphylococcus aureus | + | − | − |
| ATCC 29740 | Staphylococcus aureus | + | − | − |
| ATCC 43764 | Staphylococcus chromogenes | + | − | − |
| ATCC 29970 | Staphylococcus haemolyticus | + | − | − |
| ATCC 29062 | Staphylococcus sciuri | + | − | − |
| ATCC12228 | Staphylococcus epidermidis | + | − | − |
| ATCC 33591 | Staphylococcus aureus | + | + | − |
| ATCC 43300 | Staphylococcus aureus | + | + | − |
| ATCC 13813 | Streptococcus agalactiae | − | − | − |
| ATCC 9927 | Streptococcus uberis | − | − | − |
| ATCC 43078 | Streptococcus dysgalactiae | − | − | − |
| ATCC 4356 | Lactobacillus acidophilus | − | − | − |
| ATCC 11454 | Lactobacillus lactis | − | − | − |
| ATCC 8293 | Lactobacillus mesenteroides | − | − | − |
| ATCC 11731 | Micrococcus aurantiacus | − | − | − |
| ATCC 19433 | Enterococcus faecalis | − | − | − |
| ATCC 11563 | Aerococcus viridans | − | − | − |
| ATCC 33317 | Streptococcus bovis | − | − | − |
| ATCC 25922 | Escherichia coli | − | − | − |
| ATCC 25931 | Shigella sonnei | − | − | − |
| ATCC 70063 | Klebsiella pneumoniae | − | − | − |
The DNA from bacterial isolates were extracted from overnight grown staphylococcal isolates by QIAamp DNA minikit (Qiagen, Duesseldorf, Germany) as per manufacturer’s recommendations. The purity and concentration of the extracted DNA were tested by NanoDrop 2000c (Thermo Fischer Scientific Inc, USA). Absorbance 260 nm/280 nm ratio of ~1.8 was accepted as pure for DNA.
Precipitation was carried out to concentrate the extracted DNA. Briefly, 1/10th volume of 3 M sodium acetate (pH = 5.2 with glacial acetic acid) was added to the extracted DNA. About 2–3 volume of 100% ice cold ethanol was added and the suspension was stored at −20°C for 1 h. After centrifuging at 12,000 rpm for 15–20 min, the ethanol was decanted, and the pellet was washed with 70% ethanol. The pellet was allowed to air dry and finally resuspended in TE buffer to get concentrated DNA.
The mPCR was designed to target three genes simultaneously, namely, 16S rRNA gene for the identification of genus Staphylococcus; mecA, and mecC genes for the detection of methicillin resistance. The oligonucleotide sequences for 16S rRNA and mecA genes were obtained from previously published reports [20,21]. The oligonucleotide sequences for mecC gene were designed from the sequences available in the GenBank database (GenBank Accession No: MH448900) [Table 2]. The oligonucleotide sequences were examined for specificity and PCR suitability using the National Centre for Biotechnology Information Primer-BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Table 2: Oligonucleotide sequences used in multiplex PCR
| Gene targeted | Primer name | Oligonucleotide sequence (5’-3’) | Amplicon size (bp) | Annealing temp |
|---|---|---|---|---|
| 16S rRNA | 16S rRNA_F | GTGATCGGCCACACTGGA | 842 bp | 52°C |
| 16S rRNA_R | CAACTTAATGATGGCAACTAAGC | |||
| mecA | mecA_F | ACGAGTAGATGCTCAATATAA | 293 bp | |
| mecA_R | CTTAGTTCTTTAGCGATTGC | |||
| mecC | mecC_F | GCTCCTAATGCTAATGCA | 584 bp | |
| mecC_R | GGCTTAGAACGCCTCTATGA |
PCR: Polymerase chain reaction
The mPCR reaction mixture comprised PCR grade water, template DNA, PCR buffer, MgCl2, dNTPs, primers, Bovine serum albumin, and Taq DNA polymerase at working concentrations, as described in Table 3. The thermocycling conditions include initial denaturation at 94°C for 5 min, followed by 35 cycles with denaturation at 94°C for 1 min, annealing at 52.5°C for 1 min, and extension at 72°C for 1 min, followed by a final extension at 72°C for 10 min. The PCR amplicons were examined on 1.5% agarose gel containing ethidium bromide (10 μg/mL). A mixture of DNAs from the reference strain ATCC 33591 (positive for Staphylococcus genus and mecA) and from strain BH32 (positive for Staphylococcus genus and mecC) was used as a positive control.
Table 3: Multiplex PCR assay reaction mixture setup
| Reagent | Stock concentration | Volume (µL) | Working concentration |
|---|---|---|---|
| Sterile H2O | 11.75 | ||
| PCR Buffer | 10×(25 mM) | 2.5 | 1×(2.5 mM) |
| dNTP’s | 10 mM | 0.75 | 0.3 mM |
| MgCl2 | 25 mM | 2.5 | 2.5 mM |
| 16S rRNA (FP+RP) | 5 pmoles/µL each | 0.75*2 | 0.15 pmoles/µL each |
| mecA (FP+RP) | 5 pmoles/µL each | 0.75*2 | 0.15 pmoles/µL each |
| mecC (FP+RP) | 20 pmoles/µL | 0.75*2 | 0.6 pmoles/µL |
| Template | Variable | 2 | Variable |
| Taq DNA polymerase | 5 U/µL | 0.5 | 2.5 U |
| Bovine serum albumin | 20 mg/ml | 0.5 | 0.4 mg/ml |
| Total reaction volume | 25 |
PCR: Polymerase chain reaction, FP: Forward primer; RP: Reverse primer;
dNTP’s: Deoxynucleotide triphosphates (dNTPs: dATP, dCTP, dGTP, dTTP)
The DNA extracted from strain ATCC 33591 was spiked with the known concentration of the DNA extracted from strain BH32. Serial ten-fold dilutions equivalent to 1000 ng/mL (3.32 × 108 genome copies/mL)–0.1 ng/mL (3.32 × 104 genome copies/mL) of purified DNA was made and examined by the 16SrRNA, mecA, and mecC specific primer sets in a multiplex reaction. Each standard dilution was run in triplicates.
The mPCR specificity was determined by amplifying 10 ng/mL of DNA from Staphylococcus field isolates (n = 40), of which 32 isolates were mecA positive and 1 isolate was mecC positive). The remaining seven Staphylococcus isolates were only genus positive. None of the isolates were positive for both mecA and mecC genes. In addition, specificity of the mPCR assay was further evaluated by testing 10 ng/mL of extracted DNA from 13 ATCC non-Staphylococcus isolates and nine ATCC Staphylococcus isolates. A negative template control was incorporated in each run.
The emergence of multidrug-resistant MRSA clones poses an ongoing challenge for infection control specialists. Hence, it is important to track the epidemiology of MRSA to limit their transmission within the community [22]. The traditional methods used for epidemiological surveillance of MRSA are expensive, time-consuming, with less sensitivity, and robustness. PCR based assays are known for immense flexibility, higher efficiency, increased sensitivity, and specificity than conventional methods. The detection of methicillin resistance by PCR based methods has been described in several reports [23-25]; however, these studies targeted only mecA gene individually or in association with virulence factors like pvl [21,26]. Herein, we developed a mPCR assay for concurrent detection of genus Staphylococcus (16S rRNA gene) and its methicillin resistance determinants, namely, mecA and mecC genes [Figure 1].
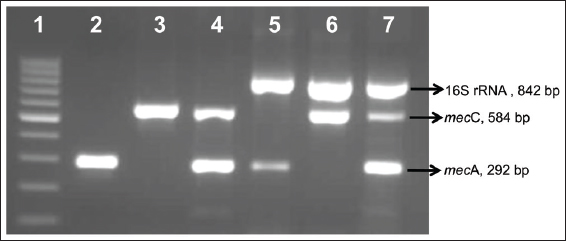 | Figure 1: Standardization of multiplex polymerase chain reaction. Lane 1: 100 bp marker; Lane 2: mecA positive; Lane 3: mecC positive; Lane 4: mecC and mecA positive; Lane 5: Staphylococcus and mecA positive; Lane 6: Staphylococcus and mecC positive; Lane 7: Staphylococcus, mecC and mecA positive [Click here to view] |
In the previous publications, misidentification of some important CoNS was reported, as these studies possibly did not aim fully conserved region in the 16S rRNA gene of Staphylococcus [23,27]. In the present study, Staphylococcus genus-specific primer targeting conserved 16S rRNA gene [15] was used which allowed to detect Staphylococcus precisely without any ambiguity. The mPCR detected 16S rRNA gene with precisely among staphylococcal strains. The detection of methicillin resistance is further complicated with the recent discovery of the mecALGA251 (mecC) gene within a novel SCCmec XI element in S. aureus [28] and CoNS [29]. In view of mecC harboring Staphylococcus spp., mecA negative results alone can no longer be a final conclusive report to exclude methicillin resistance. Genotyping methods with exclusion of mecC gene for methicillin resistance would lead to contradictory outcomes when compared to phenotypic methods [30]. Though, demonstration of mecA gene by PCR is known to be a gold standard method for the identification of methicillin resistance; however, the detection of mecC gene has become equally important in ascertaining methicillin-resistant staphylococci [31]. The strength of the present study was the incorporation of mecC specific primers in addition to the most commonly targeted mecA gene. Although, the inclusion of recently discovered methicillin-resistant determinants, i.e., mecB and mecD would have increased the significance of the study [13-15]. However, it was not practically possible for us to target these genes in view of the absence of confirmed mecB and mecD positive strains (positive controls). Considering the priority, this will be our future strategy to incorporate mecC and mecD genes as an advancement of our mPCR assay.
The analytical sensitivity of our mPCR assay was determined by testing the serial ten-fold dilutions equivalent from 1000 ng/mL (3.32 × 108 genome copies/mL) to 0.1 ng/mL (3.32 × 104 genome copies/mL) of purified DNA known to be positive for 16S rRNA, mecA, and mecC genes. The primer sets were tested in three different combinations (Staphylococcus + mecA; Staphylococcus + mecC; and Staphylococcus + mecA + mecC). The mPCR assay could detect 1 ng/ml of positive DNA, which corresponds to 1.1 × 109genome copies/ml for 16S rRNA gene, 3.17 × 109 genome copies/ml for mecA gene, and 1.6 × 109 genome copies/ml for mecC gene [Figure 2]. A study from Malaysia reported the analytical sensitivity of the pentaplex PCR to be 10 ng/mL [21]. Similarly, Rocchetti et al., [3] in 2018, showed the sensitivity up to a dilution of 10−2 for S. aureus and 10−5 for CoNS. This difference in the analytical sensitivity among the studies is inevitable, as it depends on the complexity of multiplexing, the sequences targeted, primer pair efficiency, and assay conditions.
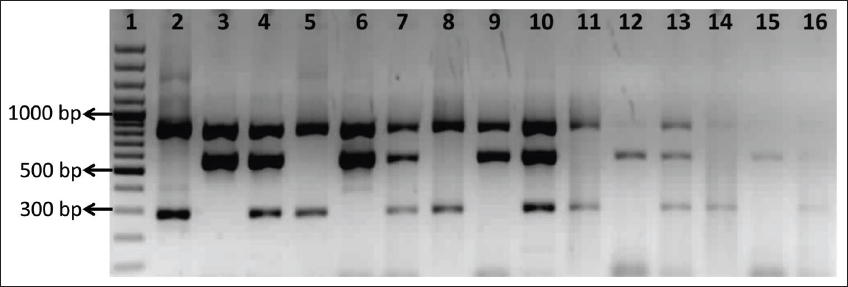 | Figure 2: Analytical sensitivity of the multiplex polymerase chain reaction. Lane 1: 100 bp ladder; Lane 2-4: 1000 ng/mL (Lane 2: Staphylococcus and mecA, Lane 3: Staphylococcus and mecC, Lane 4: Staphylococcus, mecA, and mecC); Lane 5-7: 100 ng/mL (Lane 5: Staphylococcus and mecA, Lane 6: Staphylococcus andmecC, Lane 7: Staphylococcus, mecA, and mecC); Lane 8-10: 10 ng/mL (Lane 8: Staphylococcus and mecA, Lane 9: Staphylococcus andmecC, Lane 10: Staphylococcus, mecA, and mecC); Lane 11-13: 1 ng/mL (Lane 11: Staphylococcus and mecA, Lane 12: Staphylococcus andmecC, Lane 13: Staphylococcus, mecA, and mecC); Lane 14-16: 0.1 ng/mL (Lane 14: Staphylococcus and mecA, Lane 15: Staphylococcus and mecC, Lane 16: Staphylococcus, mecA, and mecC) [Click here to view] |
The expression of mecA and mecC genes is variable and relies on several factors such as pH, temperature, media composition, inoculum size, salt concentration, and incubation time of cultures. Although, the phenotypic detection methods are widely accepted, they have their own limitations as the phenotypic expression of methicillin resistance is often heterogeneous, which describes a cell population wherein a small fraction of cells display high-level methicillin resistance [19]. All of these features highlight the necessity to develop a fast, precise, and sensitive method for the detection of methicillin resistance in staphylococci, which is independent of all these variables [32,33]. Evaluation of our mPCR assay with thirty known mecA positive and mecC negative staphylococci showed specific amplification for genus staphylococci and mecA gene among all thirty isolates tested. As expected, no amplification for mecC gene was observed among all thirty mecA positive Staphylococcus strains. Among nine ATCC Staphylococcus strains, amplification was observed with all genus specific primers while two of them, ATCC 33591 and ATCC 43300 turned positive for mecA gene as well. However, none of the non-staphylococcal ATCC reference strains (n = 13) showed amplification to any of the three genes tested. Thus, the mPCR assay exhibited 100% specificity [Table 1].
Before the discovery of mecC gene, targeting mecA gene alone was considered as criteria for the detection of methicillin resistance. Hence, duplex PCR targeting nuc gene, a species-specific marker for S. aureus and mecA gene with altered PBP2a was considered the gold standard for detection of MRSA isolates. In 2018, Rocchetti et al. [3] standardized mPCR for the identification of genus Staphylococcus and mecA gene, further differentiating S. aureus from CoNS using coa gene. Previously, Maes et al. [23] in 2002 developed a triplex PCR which included 16S rRNA, mecA and nuc gene-specific primers for the rapid characterization of staphylococci in positive blood cultures. However, the discovery of the mecC gene has challenged the microbiological laboratories and clinicians in terms of the detection of methicillin resistance and prophylaxis/treatment options. Thus, it is imperative to have a diagnostic assay targeting mecC resistant determinant as well, which will provide the exact burden of the methicillin-resistant staphylococci without any ambiguity.
Inclusion of mecA and mecC specific gene targets is an attempt to narrow down the gap in the detection of methicillin resistance. The mPCR assay was able to identify the genus Staphylococcus, methicillin resistance determinants with 100% sensitivity and specificity. This assay could be successfully used in clinical laboratory settings for rapid identification of methicillin-resistant staphylococci, which in turn may help in adopting strategies for timely and effective treatment and control options.
Authors declared that they do not have any conflicts of interest.
None.
1. Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol 2010;5:183-95. [CrossRef]
2. Ki V, Rotstein C. Bacterial skin and soft tissue infections in adults: A review of their epidemiology, pathogenesis, diagnosis, treatment and site of care. Can J Infect Dis Med Microbiol 2008;19:173-84. [CrossRef]
3. Rocchetti TT, Martins KB, Martins PY, Oliveira RA, Mondelli AL, Fortaleza CM, et al. Detection of the mecA gene and identification of Staphylococcus directly from blood culture bottles by multiplex polymerase chain reaction. Braz J Infect Dis 2018;22:99-105. [CrossRef]
4. Sawant AA, Gillespie BE, Oliver SP. Antimicrobial susceptibility of coagulase-negative Staphylococcus species isolated from bovine milk. Vet Microbiol 2009;134:73-81. [CrossRef]
5. Thorberg BM, Kuhn I, Aarestrup FM, Brandstrom B, Jonsson P, Danielsson-Tham ML. Pheno and genotyping of Staphylococcus epidermidis isolated from bovine milk and human skin. Vet Microbiol 2006;115:163-72. [CrossRef]
6. Jonas D, Speck M, Daschner FD, GrundmannH. Rapid PCR-based identification of methicillin-resistant Staphylococcus aureus from screening swabs. J Clin Microbiol 2002;40:1821-3. [CrossRef]
7. Cooper BS, Stone SP, Kibbler CC, Cookson BD, Roberts JA, Medley GF, et al. Isolation measures in the hospital management of methicillin resistant Staphylococcus aureus (MRSA): Systematic review of the literature. BMJ 2004;329:533. [CrossRef]
8. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): Guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 2009;53:4961-7. [CrossRef]
9. Barbier F, Ruppe E, Hernandez D, Lebeaux D, Francois P, Felix B, et al. Methicillin-resistant coagulase-negative staphylococci in the community: High homology of SCCmecIVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis 2010;202:270-81. [CrossRef]
10. Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden MT, et al. International working group on the classification of staphylococcal cassette chromosome elements (IWG-SCC): Guidelines for reporting novel mecA gene homologues. Antimicrob Agents Chemother 2012;56:4997-9. [CrossRef]
11. Garcia-Alvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, et al. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: A descriptive study. Lancet Infect Dis 2011;11:595-603. [CrossRef]
12. Baba T, Kuwahara-Arai K, Uchiyama I, Takeuchi F, Ito T, Hiramatsu K. Complete genome sequence of Macrococcus caseolyticus strain JCSCS5402, reflecting the ancestral genome of the human-pathogenic staphylococci. J Bacteriol 2009;191:1180-90. https://doi.org/10.1128/JB.01058-08 [CrossRef]
13. Gómez-Sanz E, Schwendener S, Thomann A, Brawand SG, Perreten V. First Staphylococcal Cassette Chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus caseolyticus isolate from a canine infection. Antimicrob Agents Chemother 2015;59:4577-83. [CrossRef]
14. Gómez-Sanz E, Schwendener S, Thomann A, Brawand SG, Perreten V. Correction for Gómez-Sanz et al. "first staphylococcal cassette chromosome mec containing a mecB-carrying gene complex independent of transposon Tn6045 in a Macrococcus caseolyticus isolate from a Canine infection". Antimicrob Agents Chemother 2018;62:e01916-18. [CrossRef]
15. Schwendener S, Cotting K, Perreten V. Novel methicillin resistance gene mecD in clinical Macrococcus caseolyticus strains from bovine and canine sources. Sci Rep 2017;7:43797. [CrossRef]
16. Brown DF, Edwards DI, Hawkey PM, Morrison D, Ridgway GL, Towner KJ, et al. Hospital infection society; infection control nurses association. Guidelines for the laboratory diagnosis and susceptibility testing of methicillin-resistant Staphylococcus aureus (MRSA). J Antimicrob Chemother 2005;56:1000-18. [CrossRef]
17. Anand KB, Agrawal P, Kumar S, Kapila K. Comparison of cefoxitin disc diffusion test, oxacillin screen agar, and PCR for mecA gene for detection of MRSA. Indian J Med Microbiol 2009;27:27-9.
18. Ferreira A, Sue D, OByrne CP, Boor KJ. Role of Listeria monocytogenes sigma(B) in survival of lethal acidic conditions and in the acquired acid tolerance response. Appl Environ Microbiol 2003;69:2692-8. [CrossRef]
19. Sakoulas G, Gold HS, Venkataraman L, De Girolami PC, Eliopoulos GM, Qian Q. Methicillin-resistant Staphylococcus aureus: Comparison of susceptibility testing methods and analysis of mecA-positive susceptible strains. J Clin Microbiol 2001;39:3946-51. [CrossRef]
20. Shome BK, Natesan K, Mitra SD, Venugopal N, Mani B, Ganaie F, et al. Development of simplex-PCR assays for accurate identification of nine staphylococcal species at genus and species levels. J Microbiol Infect D 2018;8:120-7. [CrossRef]
21. Al-Talib H, Yean CY, Al-Khateeb A, Hassan H, Singh KK, Al-Jashamy K, et al. A pentaplex PCR assay for the rapid detection of methicillin-resistant Staphylococcus aureus and panton-valentine leucocidin. BMC Microbiol 2009;9:113. [CrossRef]
22. Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. Antibiotic resistance: A rundown of a global crisis. Infect Drug Resist 2018;11:1645-58. [CrossRef]
23. Maes N, Magdalena J, Rottiers S, De Gheldre Y, Struelens MJ. Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative Staphylococci and determine methicillin resistance from blood cultures. J Clin Microbiol 2002;40:1514-7. [CrossRef]
24. Adams DN. Shortcut method for extraction of Staphylococcus aureus. DNA from blood cultures and conventional cultures for use in real-time PCR assays. J Clin Microbiol 2005;43:2932-3. [CrossRef]
25. Hogg GM, McKenna JP, Ong G. Rapid detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus directly from positive BacT/Alert blood culture bottles using real-time polymerase chain reaction: Evaluation and comparison of 4 DNA extraction methods. Diagn Microbiol Infect Dis 2008;61:446-52. [CrossRef]
26. Okolie CE, Wooldridge KG, Turner DP, Cockayne A, James R. Development of a new pentaplex real-time PCR assay for the identification of poly-microbial specimens containing Staphylococcus aureus and other staphylococci, with simultaneous detection of staphylococcal virulence and methicillin resistance markers. Mol Cell Probes 2015;29:144-50. [CrossRef]
27. Jaffe RI, Lane JD, Albury SV, Niemeyer DM. Rapid extraction from and direct identification in clinical samples of methicillin-resistant staphylococci using the PCR. J Clin Microbiol 2000;38:3407-12. [CrossRef]
28. Shore AC, Deasy EC, Slickers P, Brennan G, O'Connell B, Monecke S, et al. Detection of staphylococcal cassette chromosome mec Type 11 carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2011;55:3765-73. [CrossRef]
29. Loncaric I, Kubber-Heiss A, Posautz A, Stalder GL, Hoffmann D, Rosengarten R, et al. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J Antimicrob Chemother 2013;68:2222-5. [CrossRef]
30. Kriegeskorte A, Ballhausen B, Idelevich EA, Köck R, Friedrich AW, Karch H, et al. Human MRSA isolates with novel genetic homolog, Germany. Emerg Infect Dis 2012;18:1016-18. [CrossRef]
31. Paterson GK, Morgan FJ, Harrison EM, Peacock SJ, Parkhill J, Zadoks RN, et al. Prevalence and properties of mecC methicillin-resistant Staphylococcus aureus (MRSA) in bovine bulk tank milk in Great Britain. J Antimicrob Chemother 2014;69:598-602. [CrossRef]
32. Zheng X, Kolbert CP, Varga-Delmore P, Arruda J, Lewis M, Kolberg J, et al. Direct mecA detection from blood culture bottles by branched-DNA signal amplification. J Clin Microbiol 1999;37:4192-3. [CrossRef]
33. Martineau F, Picard FJ, Lansac N, Ménard C, Roy PH, Ouellette M, et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrob Agents Chemother 2000;44:231-8. [CrossRef]
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Year
Month