1. INTRODUCTION
Research studies revealed that the human genome encodes only 2% of protein-coding genes [1]. The pervasive transcription of the genome is leading to the generation of non-coding RNA (ncRNA) [1]. The ncRNA categorized into small ncRNA, for instance, micro-RNA (mRNA) and other class being are long ncRNAs (lncRNAs). LncRNAs are grouped into sense lncRNAs, antisense lncRNAs, intronic lncRNAs, long intergenic lncRNAs, promoter-associated lncRNAs, bidirectional lncRNAs, natural antisense transcriptions, and enhancer lncRNAs [2].
lncRNAs typically 200 nucleotides - 100 kb transcripts that lack open reading frame belongs to a team of ncRNAs [3,4]. Like, usual mRNA often codes for a protein, the lncRNAs are also spliced, capped, and polyadenylated [5]. LncRNAs possess the ability to bind to the DNA, RNA, and proteins to execute their effects on gene expression by utilizing diversified mechanisms [6].
LncRNAs regulate gene expression at transcriptional, post-transcriptional, and epigenetic levels [7]. At the epigenetic level, they regulate through RNA interference, DNA methylation or demethylation, chromatin remodeling, and histone modifications [8-14].
The lncRNAs involve in diverse functions in the cell summarized in Figure 1 [15].
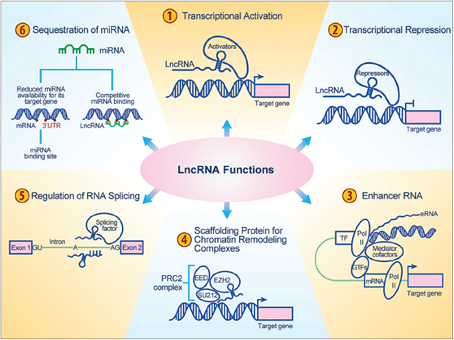 | Figure 1: Functions of long Non-coding RNAs [15] (1)Activation of target gene happens when lncRNAs interact with transcriptional activators. (2) LncRNAs represses transcription in two ways one would be keeping transcriptional activators away from chromatin and another one would be disturbing the tumour suppressor signaling. (3) The long non-coding Enhancer RNAs recruits lineage-specific complexes and regulate signaling. (4) LncRNAs recruits chromatin remodeling complexes that include PRC1 and PRC2 and serves as scaffolding proteins. (5) LncRNAs interacts with splicing factors or splicing junctions of pre-mRNA and thereby regulating the RNA splicing. (6) LncRNAs possesses binding sites for miRNAs and occupies their mRNA targets and hence serves as molecular sponges. mRNA: messenger RNA, GTF: general transcription factors, eRNA: enhancer RNA, lncRNA: long noncoding RNA, miRNA: microRNA, PRC2: polycomb repressive complex 2, UTR: untranslated region, Pol II: RNA polymerase II, TF: transcription factors. [Click here to view] |
Research studies clearly remarked that lncRNAs incredibly heterogeneous and exhibit significant functional versatility due to their conformations they assume in the course of their developmental processes and positively assisting all of them to interact with other molecules [20]. For instance, extremely diversified lncRNA interacts with various proteins which play a decisive role in the pathogenesis of various disease conditions [21].
LncRNA also experiences mutations as like protein-coding genes in such cases they become more powerful in cancers and thereby contributing to the formation of tumors and metastasis [22]. The lncRNA inactivates major tumor suppressor genes and leads to carcinogenesis [23].
LncRNAs could display characters such as promoting cancers or suppress cancers [22]. Recently, Marín-Béjar et al. [24] reported a lncRNA titled LINC-PINT (long intergenic non-protein-coding RNA, p53-induced transcript) that typically behaves as a tumor suppressor, lowering the invasive character of cancer cells. In addition, they pointed out that this is achieved merely by an interaction between highly conserved sequence elements in LINC-PINT with PRC2.
Recently, discovered lncRNAs role and mechanism of action in various cancers summarized in Table 1.
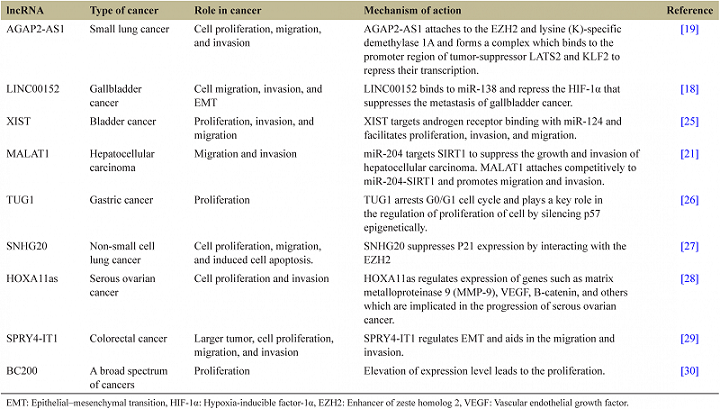 | Table 1: The lncRNAs role and mechanism of action in various cancers. [Click here to view] |
Based on some studies, lncRNAs own the potential to get translated into too short peptides; this feature is observed through ribosome protection patterns [31-33]. The regions of translated transcripts can be examined using ribosome profiling which involves the sequencing of ribosome protected fragments [31].
In yeast, Wilson and Masel [34] found that novel protein-coding genes arise from non-coding sequences. According to Xie et al., lncRNAs with active and regulated transcriptional properties could be the origin of the newly evolved proteins [35]. As reported by Ruiz-Orera et al., there are strong similarities between lncRNA molecules and recently evolved protein-coding genes [36].
Assessing the properties of the small open reading frames (sORFs) in lncRNAs and young protein-coding transcripts, Ruiz-Orera et al. found that new peptides could be encoded by lncRNAs [36]. There emerges another proof from Mackowiak et al. that sORFs get translated and exhibits some wide conservation between vertebrates and invertebrates [37].
Recently, lncRNA LINC00961 encoded a polypeptide called small regulatory polypeptide of amino acid response which aids in the muscle regeneration [38]. The data provided by Matsumoto et al., the short proteins encoded by lncRNA can orchestrate the biological processes in line with the requirements of specific tissues [38]. Another example, myoregulin a micro peptide encoded by a putative lncRNAs known to regulate skeletal muscle physiology [39].
Aggregated data shed light on the possibility of coding potential of lncRNAs according to necessities in the cell.
In case of cancer, proteins translated from lncRNAs still to be found and consequently their role in the cancers. As stated by Ruiz-Orera et al., short peptides obtained from lncRNAs undergo degradation as soon as they get translated [36]. This is the key reason which is making it so challenging to uncover the proteins encoded by lncRNAs in cancers. If this is the fact that they are synthesized in cancers, there could be a potential chance of targeting cancer.
2. CONCLUDING REMARKS
lncRNA research is a quite challenging area to study. Applying them to the significantly larger segments of the diseases and studying the role of lncRNA within the cells is in fact, intriguing.
At present, lncRNAs are in the discovery stage and their interventions in diseases. The lncRNAs are required to be evaluated individually to acquire knowledge of the precise function. One strategy could be using the drugs in combination in which one would suppress the expression of lncRNA and another drug effectively targets tumor cell [40]. Since lncRNA works effectively utilizing diverse structures, molecules that could bind to them or alters the structure, for neutralization could be developed [40].
The investigation of lncRNAs should head toward finding their role in escalation or suppression of a disease. The emergence of advanced technologies in RNA Biology made a significant pathway to find biomarkers.
3. REFERENCES
1. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Science 2005;309:1559-63.
2. Hauptman N, Glava? D., Long non-coding RNAs in cancer: Diagnostic and prognostic value. Med Chem (Los Angeles) 2016;6:422-4.
3. Mattick JS. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Reports 2001;2:986-91.
4. Lee C, Kikyo N. Strategies to identify long noncoding RNAs involved in gene regulation. Cell Biosci 2012;2:37.
5. Quinn JJ, Chang HY. Unique features of long noncoding RNA biogenesis and function. Nat Rev Gen 2016;17:47-62.
6. Parasramka MA, Maji S, Matsuda A, Yan IK, Patel T. Long non-coding RNAs as novel targets for therapy in hepatocellular carcinoma. Pharmacol Ther 2016;161:67-78.
7. Gong Z, Zhang S, Zhang W, Huang H, Li Q, Deng H, et al. Long non-coding RNAs in cancer. Sci China Life Sci 2012;55:1120-4.
8. Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 2010;38:662-74.
9. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129:1311-23.
10. Liang X, Ma J, Schatten H, Sun Q. Epigenetic changes associated with oocyte aging. Sci China Life Sci 2012;55:670-6.
11. Huang H, Jiao R. Roles of chromatin assembly factor 1 in the epigenetic control of chromatin plasticity. Sci China Life Sci 2012;55:15-9.
12. Chao S, Li J, Jin X, Tang H, Wang G, Gao G, et al. Epigenetic reprogramming of embryos derived from sperm frozen at -20 degrees C. Sci China Life Sci 2012;55:349-57.
13. Bi X. Functions of chromatin remodeling factors in heterochromatin formation and maintenance. Sci China Life Sci 2012;55:89-96.
14. Ling HY, Hu B, Feng SD, Liao DF, Wen GB. The role of epigenetic regulation in diabetes and its complications. Prog Biochem Biophys 2012;39:14-21.
15. Malik B, Feng FY. Long noncoding RNAs in prostate cancer: Overview and clinical implications. Asian J Androl 2016;18:568-74.
16. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6.
17. Hon CC, Ramilowski JA, Harshbarger J, Bertin N. An atlas of human long non-coding RNAs with accurate 5’ ends. Nature 2017;543:199-204.
18. Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li C, et al. Long non-coding RNA LINC00152 promotes gallbladder cancer metastasis and epithelial-mesenchymal transition by regulating HIF-1a via miR-138. Open Biol 2017;7:160247.
19. Li W, Sun M, Zang C, Ma P, He J, Zhang M, Shu Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis 2017;7:e2225.
20. Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Gen Biol 2017;18:206.
21. Hou Z, Xu X, Zhou L, Fu X, Tao S, Zhou J, et al. The long non-coding RNA MALAT1 promotes the migration and invasion of hepatocellular carcinoma by sponging miR-204 and releasing SIRT1. Tumor Biol 2017;39:1010428317718135.
22. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and Cancer: A new paradigm. Cancer Res 2017;77:1-17.
23. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38.
24. Marín-Béjar O, Mas AM, González J, Martinez D, Athie A, Morales X. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol 2017;18:202.
25. Xiong Y, Wang L, Li Y, Chen M, He W, Qi L. The long non-coding RNA XIST interacted with MiR-124 to modulate bladder cancer growth, invasion, and migration by targeting androgen receptor (AR). Cell Physiol Biochem 2017;43:405-18.
26. Zhang E, He X, Yin D, Han L, Qiu M, Xu T, et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis 2017;7:e2109.
27. Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B, et al. Long non-coding RNA SNHG20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis 2017;8:e3092.
28. Yim GW, Kim HJ, Kim LK, Kim SW, Kim S, Nam EJ, et al. Long non-coding RNA HOXA11 antisense promotes cell proliferation and invasion and predicts patient prognosis in serous ovarian cancer. Cancer Res Treat Off J Korean Cancer Assoc 2017;49:656.
29. Shen F, Cai WS, Feng Z, Chen JW, Feng JH, Liu QC, et al. Long non-coding RNA SPRY4-IT1 pormotes colorectal cancer metastasis by regulate epithelial-mesenchymal transition. Oncotarget 2017;8:14479.
30. Booy EP, McRae EK, Koul A, Lin F, McKenna SA. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer 2017;16:109.
31. Ingolia NT. Ribosome profiling: New views of translation, from single codons to genome scale. Nat Rev Genet 2014;15:205-13.
32. Bazzini AA, Johnstone TG, Christiano R, Mackowiak SD, Obermayer B, Fleming ES, et al. Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J 2014;33:981-93.
33. Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in arabidopsis. Proc Natl Acad Sci U S A 2014;111:E203-12.
34. Wilson BA, Masel J. Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol Evol 2011;3:1245-52.
35. Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, et al. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet 2012;8:e1002942.
36. Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM. Long non-coding RNAs as a source of new peptides. Elife 2014;3:E03523.
37. Mackowiak SD, Zauber H, Bielow C, Thiel D, Kutz K, Calviello L, et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol 2015;16:179.
38. Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, et al. MTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 2017;541:228-32.
39. Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 2015;160:595-606.
40. Hauptman N, Glava? D. Long non-coding RNA in cancer. Int J Mol Sci 2013;14:4655-69.