1. INTRODUCTION
The cultivated strawberry (Fragaria × ananassa Duch.) is one of the most popular species of the Rosaceae family, due to its nutritional importance, aroma, and flavor; these qualities make strawberries one of the most economically important crops [1,2]. However, the crop suffers from various fungal diseases, such as Colletotrichum acutatum, which causes serious economic losses to strawberry production as a result of the damage caused both on immature fruit before harvest and on ripe fruit at harvest or in the post-harvest storage stage [3].
In the search for genes related to resistance and susceptibility to fungal diseases such as those caused by C. acutatum, a small family of peptides has been identified, which are the Rapid alkalinization factors (RALFs), which also encode ubiquitous small peptides that stimulate apoplastic alkalinization through interaction with the receptor kinase and act as negative regulators of the plant’s immune response [4]. Likewise, Rapid alkalinization factor 33 (RALF33) was found to play a key role in the regulation of plant defense and development [4,5]. This was demonstrated by silencing the FaRALF33 gene in red fruits inoculated with C. acutatum, reducing pathogen infection in the fruit, while overexpression of the gene decreased fruit resistance to the fungus [5].
Gene inactivation is currently being developed by precision gene editing in the genome through the clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) system [6]. This system features two main components: Cas9 endonuclease to introduce double-strand breaks (DSBs) at a specific genomic site, and single guide RNA (sgRNA), which is a sequence complementary to the target DNA sequence [7,8]. DSBs caused by the sgRNA/Cas9 complex are repaired by the endogenous non-homologous end joining (NHEJ) mechanism [9], which is error-prone and often generates small deletions or insertions [10,11]. These null alleles or coding regions of the genome that arose by NHEJ repair mechanism trigger premature stop codons (frameshift mutation), resulting in loss of function or knockout [12]. Due to its simplicity, versatility and efficiency, CRISPR/Cas technology has successfully edited the genome of globally agronomically important species [13].
It is important to note that to develop knockouts using the CRISPR/Cas9 system, for example, in plant susceptibility genes, the design of more than one sgRNA is necessary to increase the success rate and previously this is developed in silico [14,15]. There are numerous online bioinformatics tools accessible to design of these sgRNAs, such as CHOPCHOP, CRISPRlnc, CRISPR RGEN tools, and sgRNA scorer 2.0 [16,17]. All these tools and in silico designs allow efficient design of sgRNAs. This is of great importance as a starting point and fundamental in any gene editing process.
The present study aims to design in silico sgRNAs for CRISPR/Cas9-mediated RALF33 gene mutagenesis in strawberry (F. × ananassa Duch.), which would lead to a decrease in the infection process of C. acutatum.
2. MATERIALS AND METHODS
2.1. FaRALF33 Peptide Sequence Identification and Alignment
For the identification of the FaRALF33 gene sequence, the NCBI database [https://www.ncbi.nlm.nih.gov/] was searched for the RALF33 gene in Fragaria vesca. The RALF33 gene (gene ID: 101302223) with location NC_020493.1 (20956047-20959174/LG2) and accession code XM_011460413.1 was selected. From the FvRALF33 sequence, the sequence prediction of FaRALF33 was started by aligning the first sequence with the genome of F. × ananassa cv. Wongyo 3115 (GCA_019022445.1) using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The homologous sequence in accession CM032302 and region 18337124-18337860 was selected. In addition, the RALF33 gene was selected from other plant species [Table 1] for the multiple alignments of amino acid sequences in Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). From these sequences, the phylogenetic tree was constructed by neighbor-joining using the MEGA X software (https://www.megasoftware.net/), with a fixed value in 1000 bootstrap.
Table 1: RALF33 gene accessions from plant species selected for multiple amino acid sequence alignment for the FaRALF33 gene.
| S. No. | Accesión | Specie | Gene | Percent identity |
|---|---|---|---|---|
| 1. | XP_011458715.1 | Fragaria vesca subspp. vesca | FvRALF33 | 100.00 |
| 2. | XP_050363374.1 | Potentilla anserina | PaRALF33 | 96.49 |
| 3. | XP_024165362.1 | Rosa chinensis | RchRALF33 | 96.49 |
| 4. | XP_007199089.2 | Prunus persica | PpRALF33 | 76.07 |
| 5. | XP_002531878.1 | Ricinus communis | RcRALF33 | 72.65 |
| 6. | XP_016709980.2 | Gossypium hirsutum | GhRALF33 | 76.15 |
| 7. | XP_006444286.1 | Citrus clementina | CcRALF33 | 74.07 |
| 8. | XP_008386646.2 | Malus domestica | MdRALF33 | 68.83 |
| 9. | XP_007050868.2 | Theobroma cacao | TcRALF33 | 72.73 |
| 10. | XP_039012113.1 | Hibiscus syriacus | HsRALF33 | 68.70 |
| 11. | NP_567476.1 | Arabidopsis thaliana | AtRALF33 | 64.91 |
2.2. In silico Design of sgRNAs for FaRALF33 using a CRISPR/Cas9 Approach
The design of the sgRNAs for the target region of the FaRALF33 gene was performed using the online tool CHOPCHOP (https://chopchop.cbu.uib.no/) on the F. × ananassa (FaRR1) genome. In this process, the CRISPR/Cas9 function for knock-out was used to perform recognition and frameshift mutation in the on-target sequence. Suitable sequences were selected and evaluated according to the criteria of GC content between 35% and 65% [18], efficiency ≥40, and self-complementarity <1. Suitable sgRNA sequences were discriminated by Mismatches (MM) from 0 to 3, with the aim of not obtaining or reducing off-target sequences.
2.3. In silico Prediction of sgRNA Structure for FaRALF33
For the prediction of the structures of the selected gRNAs, the RNA scaffold sequence (5’-GUUUUUUAGAGA GCUAGAAAAAUAGCAAGCAAGUUAAAAUAAGG CUAGUCCGUUAUCAACUUGAAAAAGUGGCACCGAGUCG GUGCUUUUUU-3’) was added, and the secondary structure was obtained using the online tool RNA fold webserver belonging to The Vienna RNA Web Services (http://rna.tbi.univie.ac.at/#webservices). For the discrimination of the selected sgRNAs, their secondary structure was evaluated, and the formation of one to two hairpins at the upper end, at the lower end (stem) the presence of 2–3 loops within the structure was established.
2.4. In silico Design of a Vector for CRISPR/Cas9-Mediated Mutagenesis of the FaRALF33 Gene
For the in silico design of the FaRALF33 gene mutagenesis vector, the free software A plasmid Editor was used, using the pCas9-TPC vector (https://www.addgene.org/61478/) as a base. The vector was modified by inserting the plant marker green fluorescent protein (GFP) and sgRNAs into the multiple cloning site, GFP was inserted into the SmaI restriction site, while the sgRNAs together with the Arabidopsis thaliana U6-26 promoter (AtU6-26) were inserted into the MluI and PacI restriction sites for sgRNA 1, while sgRNA 2 was inserted into the SpeI and XbaI restriction sites.
3. RESULTS AND DISCUSSION
3.1. FaRALF33 Peptide Sequence Identification and Alignment
The predicted sequence of the FaRALF33 gene has a length of 737 bp, while the coding sequence (CDS) has a length of 345 bp coding for a peptide of 114 amino acids [Figure 1], with a coding length of 100% similarity to the RALF33 gene in F. vesca [Table 1], compared to the length of the FvRALF33 gene (735), it has two gaps at position 140 of its sequence [5]. Multiple alignment of homologous RALF33 sequences from other plant species showed that the amino acid sequence of FaRALF33 features typical sequences of known RALF peptides, as is the RRILA proteolytic cleavage site for propeptide processing by subtilisin-like protease and release of the mature peptide [Figure 2a]. The FaRALF33 sequence by presenting the YISY, YYNC, and RCRR/RCRS motifs in its mature form, complies with the characteristics of the RALF peptide clade I subfamily [19]. Within the RALF33 homolog sequence cluster, the close grouping of FaRALF33 with FvRALF33, followed by RcRALF33 and PaRALF33 can be evidenced, indicating that RALF33 in strawberries shares a close relationship with those of roses and silverweed, whereas, with cotton, cocoa, clementine, and A. thaliana it presents a distant relationship, the latter species being the most distant [Figure 2b]. This distance could be related to the function of AtRALF33, which has been determined as a peptide that regulates seedling growth and has a greater effect on root growth inhibition due to severe compression of the meristematic and elongation zones [20].
 | Figure 1: Rapid alkalinization factors 33 sequence of Fragaria × ananassa (FaRALF33) cultivar Wongyo, gene sequence (yellow), CDS sequence (green) and peptide sequence (turquoise). [Click here to view] |
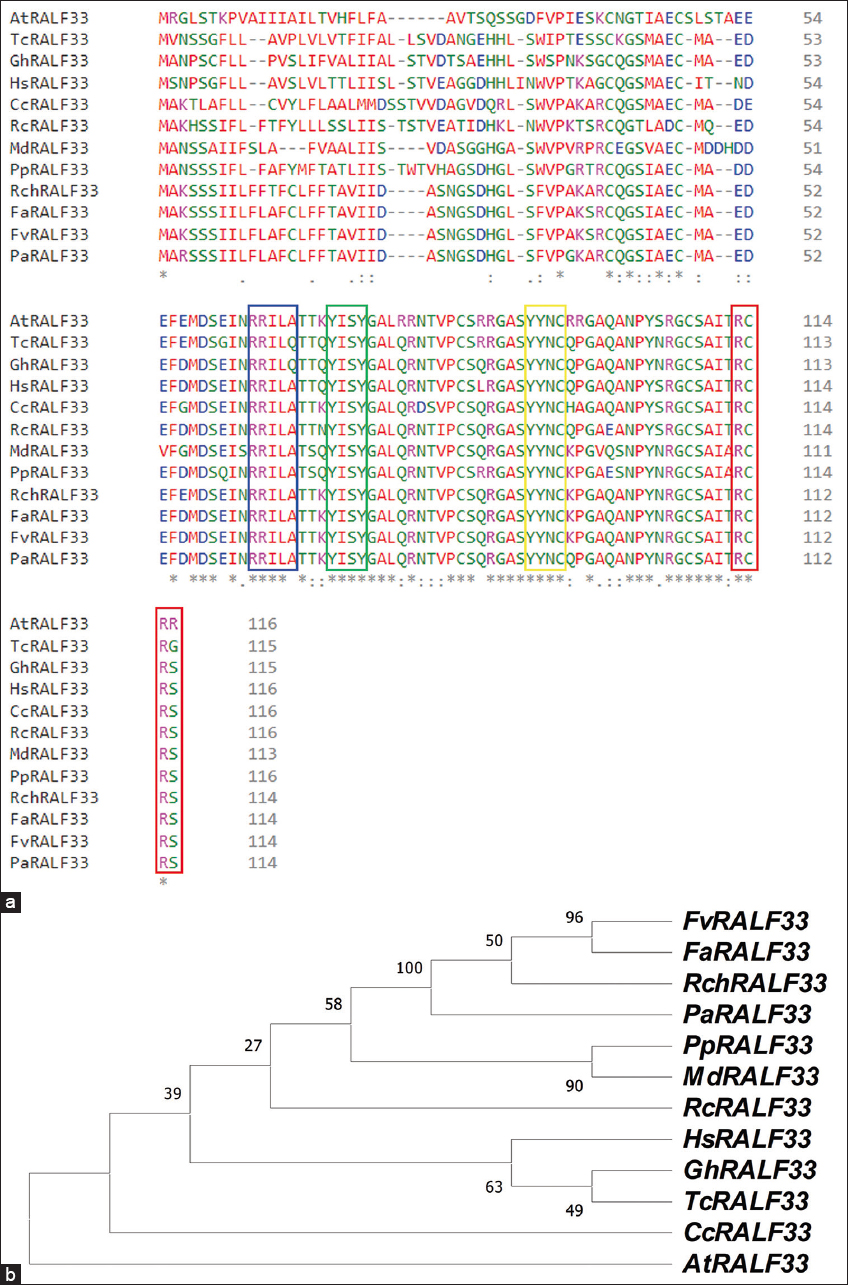 | Figure 2: Sequence of rapid alkalinization factor 33 (RALF33) peptide from Fragaria × ananassa compared with RALF33 peptide sequence from other plant species. (a) Multiple sequence alignment of RALF33 peptide sequences and their typical sequences of known RALF peptides: the proteolytic cleavage site (RRILA) is enclosed in a light blue box, the YISY motif is enclosed in a green box, the YYNC motif is enclosed in a yellow box, and the RCRR/RCRS motif is enclosed in a red box. (b) Phylogenetic tree analysis of FaRALF33 and RALF33 peptide sequences from other selected species. [Click here to view] |
3.2. In silico Design of sgRNA for FaRALF33
In the in silico design of the sgRNAs, a total of 73 candidate sgRNA sequences were found from which 19 top hits were selected and categorized as pre-selected sequences [Table 2]. To determine the pre-selected sequences, GC content was considered, selecting sequences that presented ranges of 35 to 65 GC%, genomic editing efficiency ≥40 and self-complementarity <1, ranges established for the selection of sgRNA top hits by Pratami et al. [18], in relation to the decrease of large number of candidate sequences. The sgRNA efficiency was calculated by “G20,” which is used by default in CHOPCHOP platform, which ranges from 0 to 100, where 100 is indicative of a successful knockout. The “G20” efficiency prioritizes guanine at position 20, just after PAM [21].
Table 2: Top hit sgRNA sequences for genomic editing in the FaRALF33 gene region.
| Target sequence | Genomic location | Strand | GC content (%) | Self-complementarity | Mismatch | Efficiency | ||
|---|---|---|---|---|---|---|---|---|
| MM0 | MM1 | MM2 | ||||||
| TTTTGGCAAATAGAGGAATGTGG | 136 | − | 35 | 0 | 1 | 0 | 1 | 57.77 |
| GCAAGCAACGGCAGCGATCACGG | 258 | + | 60 | 1 | 2 | 0 | 0 | 43.75 |
| ACTGTGCCATGCTCGCAAAGGGG | 426 | + | 55 | 0 | 1 | 0 | 3 | 72.81 |
| ACACTGTGCCATGCTCGCAAAGG | 424 | + | 55 | 0 | 1 | 0 | 3 | 51.28 |
| CACTGTGCCATGCTCGCAAAGGG | 425 | + | 55 | 0 | 1 | 0 | 3 | 44.64 |
| TGAATAAGCTAACCTCTGACAGG | 44 | − | 40 | 1 | 1 | 1 | 2 | 52.41 |
| CGACTCTCCCATCTTGGACTTGG | 87 | − | 55 | 0 | 3 | 0 | 0 | 45.08 |
| TGCAATGGAACCCTGGCAACGGG | 304 | − | 55 | 0 | 3 | 0 | 0 | 40.16 |
| TCAGCTATGGAGCTCTACAGAGG | 400 | + | 50 | 1 | 1 | 3 | 0 | 66.13 |
| GAAGCACCCCTTTGCGAGCATGG | 432 | − | 60 | 0 | 1 | 3 | 0 | 52.09 |
| GTAGGGGTTAGCCTGTGCCCCGG | 472 | − | 65 | 1 | 2 | 2 | 0 | 47.52 |
| GGAACCCTGGCAACGGGACTTGG | 298 | − | 65 | 0 | 3 | 1 | 0 | 41.61 |
| GTTGCTTGCATCGATGATCACGG | 244 | − | 45 | 0 | 4 | 0 | 0 | 61.2 |
| CATTGCAGAGTGCATGGCTGAGG | 320 | + | 55 | 1 | 4 | 0 | 0 | 55.65 |
| TGCACTCTGCAATGGAACCCTGG | 311 | − | 55 | 0 | 4 | 0 | 0 | 53.43 |
| CAAGCAACGGCAGCGATCACGGG | 259 | + | 60 | 1 | 4 | 0 | 0 | 51.94 |
| GAGAATTCACACATGATCTAAGG | 571 | − | 35 | 0 | 4 | 0 | 0 | 49.95 |
| CTACTACAACTGCAAGCCCGGGG | 455 | + | 55 | 1 | 3 | 1 | 3 | 69.82 |
| TCCTACTACAACTGCAAGCCCGG | 453 | + | 50 | 0 | 3 | 4 | 0 | 44.92 |
MM: Mismatches, sgRNA: Single guide RNA. Sequences and values in bold correspond to the final two sgRNAs selected
From the 19 pre-selected sequences, an additional filtration was performed, adding unwanted targets in the genome, establishing as discriminant the presence of off-targets maximum 3 for the MM0 target site, and <2 for the MM1 and MM2 targets. Four sgRNA sequences were selected from the filtration: the sequences sgRNA 1 (5’-CGACTCTCCCATCTCTTGGACT-3’), sgRNA 2 (5’-GCAAGCAACGGCAACGGCAGCGATCA-3’), sgRNA 3 (5’-TTTTTTGGCAAATAGAGAGGAATG-3’) and sgRNA 4 (5’-TGAATAAGCTAACCTCTGAC-3’), The sequences were also the main sequences recommended by the CHOPCHOP online tool, as well as the statistics of the repair profile predictions [Table 3] and the specific primers [Table 4].
Table 3: Statistics of repair profile predictions of sgRNA 1 and sgRNA 2 sequences for FaRALF33 gene editing.
| Repair profile prediction | Target sequence | |
|---|---|---|
| sgRNA 1 | sgRNA 2 | |
| Frameshift frequency | 18.15 | 52.08 |
| Precision score | 0.74 | 0.48 |
| Frame+0 frequency | 81.85 | 47.92 |
| Frame+1 frequency | 9.54 | 25.33 |
| Frame+2 frequency | 8.61 | 26.75 |
| 1-bp ins frequency | 2.86 | 11.83 |
| Highest deletion frequency | 72.03 | 20.62 |
| Highest insertion frequency | 1.30 | 6.16 |
| Highest outcome frequency | 72.03 | 20.62 |
| Microhomology deletion frequency | 88.23 | 68.78 |
| Microhomology-less deletion frequency | 8.91 | 19.39 |
sgRNA: Single guide RNA
Table 4: Sequence-specific primers sgRNA 1 and sgRNA 2 for gene editing of FaRALF33.
| Characteristics of primers | Target sequence | |
|---|---|---|
| sgRNA 1 | sgRNA 2 | |
| Left primer (5’- 3’) | AAATGAGAGCTTCCAATTCCCT | TATTCTCTTCTTGGCCTTCTGC |
| Left primer coordinates | Sequence: 7–29 | Sequence: 209–231 |
| Left primer Tm | 60.4 | 60.0 |
| Right primer (5’- 3’) | ATTTGCTTCTGGTATGCCTTGT | CAGTTGTAGTAGGAAGCACCCC |
| Right primer coordinates | Sequence: 169–191 | Sequence: 445–467 |
| Right primer Tm | 60.0 | 60.1 |
| Product size (bp) | 184 | 258 |
Tm: Melting temperature, sgRNA: single guide RNA
In the final filtering, the sequences that will present 0 for the MM1 and MM2 targets were considered, discarding the sgRNA 3 and sgRNA 4 sequences, additionally, this last sequence contains poly-T and can act as a termination signal for RNA polymerase III, so it is recommended that this poly-T be avoided in the design. Thus, sgRNA 1 and sgRNA 2, with 55% and 60% GC content, respectively, comply with one of the recommendations established for sgRNA design, being within the most effective range established by [15,22], who determine that the greatest effectiveness that a sgRNA can present is related to the GC content in ranges of 40–70% because these contents in the sgRNA sequence allow a greater interaction with Cas9 endonuclease, which leads to better activity and efficiency. The most important point of sgRNA 1 and sgRNA 2 is their target location, located firstly in the coding region and secondly located close to the N-terminal end; this is because the activity and efficiency of sgRNAs are negatively affected when they have targets close to the C-terminal end, because the mutation that can be performed at that site has a very low probability of changing the reading frame, i.e., there would be no interruption of protein expression or inactivation of the gene of interest, in addition to the fact that sgRNAs targeting non-coding regions are inefficient in the search for gene inactivation mutations [21,23].
3.3. In silico Prediction of sgRNA Secondary Structure for FaRALF33
The secondary structure of the four sgRNAs was predicted in conjunction with RNA scaffold sequence; the criteria considered were the formation of two hairpins at the upper end and an extended stem-loop (stem-loop RAR) at the lower end [23,24] [Figure 3]. These secondary structures are important in many biological processes since they can determine the accessibility of nucleotides and the resulting interactions at each locus, and they are also necessary to know since they allow the discrimination of sgRNA sequences that may present alterations and variations in their secondary structure that lead to the inefficiency of the recognition and editing process of the target region, as well as the interaction with the Cas9 protein [23]. Of the total sgRNAs selected, only three (sgRNA 1, sgRNA 2, and sgRNA 4) presented favorable structures, being sgRNA 1 and sgRNA 2 the most favored, due to the lower presence of paired seed regions; on the other hand, the presence of the first stem-loop is not necessary because 82% of sgRNAs lose in plants and it is not related to editing efficiency [24]. In the secondary structure of sgRNA 3 lacks the essential structures such as the required RAR stem-loop, second stem-loop, and third stem-loop, which gives key structural properties for processing and binding to Cas9 protein [25].
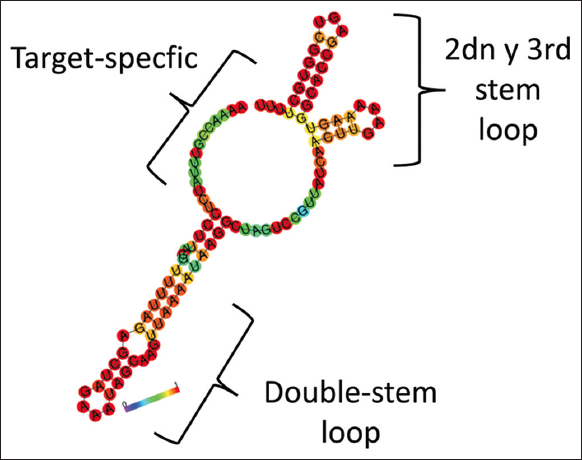 | Figure 3: Predicted secondary structure of the predicted single-guide RNA binding to the RNA scaffold sequence for mutagenesis of the FaRALF33 gene. [Click here to view] |
On the other hand, it is considered that the variation of some nucleotides determines the efficiency of sgRNAs, being significant in comparison with inefficient sgRNAs, but within these possible variations it has been established that a conserved motif is present in the sgRNA, which determines the formation of stable hairpin structures in established positions [26], allowing to obtain results in the structures of the sgRNAs selected in this work, which are in accordance with the efficient secondary structures. The prediction of secondary structures is essential to determine certain interactions resulting in various biological processes, being recognized as an important feature in the design of sgRNAs [27-29].
3.4. In silico Design of a Vector for CRISPR/Cas9-Mediated Mutagenesis of the FaRALF33 Gene
For the design of the FaRALF33 gene mutagenesis vector, the sgRNA 1 and sgRNA 2 sequences were inserted, each with their respective sRNA scaffold and driven by the AtU6-26 promoter, and additionally, the GFP marker (inserted with a 35S promoter and terminator) was added, obtaining the pCas9-TPC-GFP-2XsgRNA vector [Figure 4].
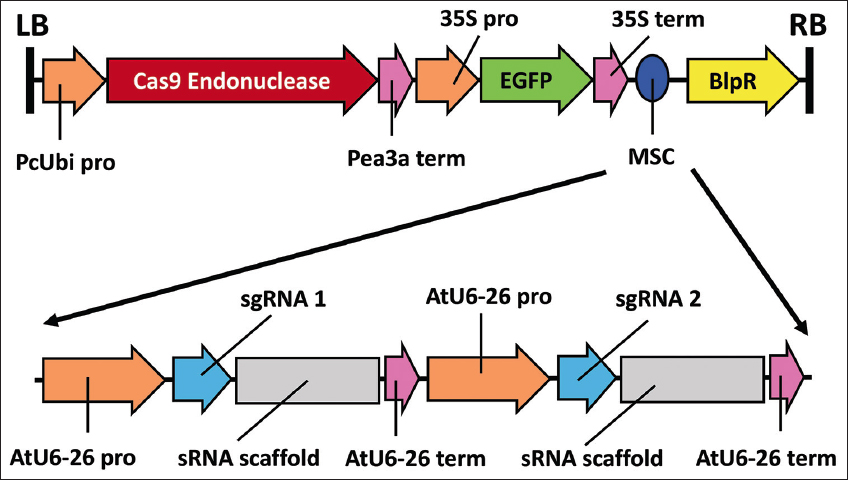 | Figure 4: T-DNA region of the pCas9-TPC-GFP-2XsgRNA vector for FaRALF33 gene mutagenesis, with two single-guide RNAs inserted into the multiple cloning site. RB and LB indicate the right border and left border of the T-DNA of the pCas9-TPC-GFP-2XsgRNA vector. [Click here to view] |
The pCas9-TPC-GFP-2XsgRNA vector was designed from the pCas9-TPC base vector that harbors the Cas9 sequence driven by Petroselinum crispum ubiquitin promoter because this promoter presents high activity and durability with the ability to maintain these characteristics in different explants under in vitro conditions [30] Thus, Poudel [31] developed the construction of the pCas9-TPC vector::F. vesca Cell Death Inducer Protein 1 (FvCDIP1)_2XgRNA, presenting as a base the same vector as presented in this work, the vector they developed resulted successful because the designed sgRNAs and the Ca9 endonuclease achieved the established objectives with the gene knockout of the FvCDIP1 gene. Likewise, demonstrating that the pCas9-TPC vector can be modified for the insertion of two sgRNA sequences because the vector was originally designed and worked with a single sgRNA [32].
The choice of the AtU6-26 promoter was made despite the recommendation in the use of native promoters, with which it is possible to obtain greater mutation efficiency in the target region [33], as was also demonstrated in strawberry, through the use of the F. vesca U6III (FvU6III) native promoter, which was more effective in obtaining mutants in F. vesca cv. Hawaii 4, in an experimental comparison of relative mutation efficiency with the A. thaliana U6-26 promoter; but in the same experiment executed in F. × ananassa cv. Calypso they obtained higher mutation efficiency with the AtU6-26 promoter than with the FvU6III promoter [34].
4. CONCLUSION
Two sgRNAs were designed in silico for CRISPR/Cas9-mediated mutagenesis of the FaRALF33 gene, with the prospect of reducing the infection process of C. acutatum. The sequence of FaRALF33 was analyzed and compared with other plant species and presented the main features of RALF peptide, presenting the YISY, YYNC, and RCRR/RCRS motifs. The predicted secondary structures of the selected sgRNAs present efficient sgRNA structures in targeted mutagenesis. Likewise, the vector pCas9-TPC-GFP-2XsgRNA, a vector with the two designed sgRNA sequences driven by the AtU6-26 promoter, was designed. This targeted mutagenesis vector was designed using in silico computational tools, as were the sgRNA and secondary structure predictions. These tools are important in predicting the accuracy and success of target target mutation, through this approach allows molecular studies and genetic engineering to leverage bioinformatics and computational techniques to practically design and predict DNA molecules or proteins.
5. AUTHORS’ CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors (ICMJE) requirements/guidelines.
6. FUNDING
There is no funding to report.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
This study does not involve experiments on animals or human subjects.
9. DATA AVAILABILITY
All the data is available with the authors and shall be provided upon request.
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Tapia RR, Barbey CR, Chandra S, Folta KM, Whitaker VM, LeeS. Evolution of the MLO gene families in octoploid strawberry (Fragaria ×ananassa) and progenitor diploid species identified potential genes for strawberry powdery mildew resistance. Hortic Res 2021;8:1-17. [CrossRef]
2. López-Casado G, Sánchez-Raya C, Ric-Varas PD, Paniagua C, Blanco-Portales R, Muñoz-Blanco J, et al. CRISPR/Cas9 editing of the polygalacturonase FaPG1 gene improves strawberry fruit firmness. Hortic Res 2023;10:uhad011. [CrossRef]
3. Prusky D. Pathogen quiescence in postharvest diseases. Annu Rev Phytopathol 1996;34:413-34. [CrossRef]
4. Guidarelli M, Carbone F, Mourgues F, Perrotta G, Rosati C, Bertolini P, et al. Colletotrichum acutatum interactions with unripe and ripe strawberry fruits and differential responses at histological and transcriptional levels. Plant Pathol 2011;60:685-97. [CrossRef]
5. Merino MC, Guidarelli M, Negrini F, De Biase D, Pession A, Baraldi E. Induced expression of the Fragaria ×ananassa rapid alkalinization factor-33-like gene decreases anthracnose ontogenic resistance of unripe strawberry fruit stages. Mol Plant Pathol 2019;20:1252-63. [CrossRef]
6. Jung C, Capistrano-Gossmann G, Braatz J, Sashidhar N, Melzer S. Recent developments in genome editing and applications in plant breeding. Plant Breed 2018;137:1-9. [CrossRef]
7. Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V. Plant genome editing made easy:Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013;9:39. [CrossRef]
8. Carneiro P, de Freitas MV, Matte U. In silico analysis of potential off-target sites to gene editing for Mucopolysaccharidosis type I using the CRISPR/Cas9 system:Implications for population-specific treatments. PLoS One 2022;17:e0262299. [CrossRef]
9. González MN, Massa GA, Andersson M, Décima Oneto CA, Turesson H, Storani L, et al. Comparative potato genome editing:Agrobacterium tumefaciens-mediated transformation and protoplasts transfection delivery of CRISPR/Cas9 components directed to StPPO2 gene. Plant Cell Tissue Organ Cult 2021;145:291-305. [CrossRef]
10. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819-23. [CrossRef]
11. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014;157:1262-78. [CrossRef]
12. García-Tuñón I, Alonso-Pérez V, Vuelta E, Pérez-Ramos S, Herrero M, Méndez L, et al. Splice donor site sgRNAs enhance CRISPR/Cas9-mediated knockout efficiency. PLoS One 2019;14:e0216674. [CrossRef]
13. Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 2013;41:e188. [CrossRef]
14. Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 2013;31:839-43. [CrossRef]
15. Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 2015;33:187-97. [CrossRef]
16. Liu X, Yang J, Song Y, Zhang X, Wang X, Wang Z. Effects of sgRNA length and number on gene editing efficiency and predicted mutations generated in rice. Crop J 2022;10:577-81. [CrossRef]
17. Xu Y, Zhang L, Lu L, Liu J, Yi H, Wu J. An efficient CRISPR/Cas9 system for simultaneous editing two target sites in Fortunella hindsii. Hortic Res 2022;9:uhac064. [CrossRef]
18. Pratami MP, Fendiyanto MH, Satrio RD, Nikmah IA, Awwanah M, Farah N, et al. In-silico genome editing identification and functional protein change of Chlamydomonas reinhardtii acetyl-CoA carboxylase (CrACCase). Jordan J Biol Sci 2022;15:431-40. [CrossRef]
19. Campbell L, Turner SR. A comprehensive analysis of RALF proteins in green plants suggests there are two distinct functional groups. Front Plant Sci 2017;8:37. [CrossRef]
20. Gjetting SK, Mahmood K, Shabala L, Kristensen A, Shabala S, Palmgren M, et al. Evidence for multiple receptors mediating RALF-triggered Ca2+signaling and proton pump inhibition. Plant J Cell Mol Biol 2020;104:433-46. [CrossRef]
21. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184-91. [CrossRef]
22. Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014;343:80-4. [CrossRef]
23. Konstantakos V, Nentidis A, Krithara A, Paliouras G. CRISPR-Cas9 gRNA efficiency prediction:An overview of predictive tools and the role of deep learning. Nucleic Acids Res 2022;50:3616-37. [CrossRef]
24. Liang G, Zhang H, Lou D, Yu D. Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci Rep 2016;6:21451. [CrossRef]
25. Hassan MM, Chowdhury AK, Islam MT. In silico analysis of gRNA secondary structure to predict its efficacy for plant genome editing. Springer Protoc Handb 2021;30:15-22. [CrossRef]
26. Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014;156:935-49. [CrossRef]
27. Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci U S A 2005;102:4006-9. [CrossRef]
28. Wong N, Liu W, Wang X. WU-CRISPR:Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol 2015;16:218. [CrossRef]
29. Xu H, Xiao T, Chen CH, Li W, Meyer CA, Wu Q, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res 2015;25:1147-57. [CrossRef]
30. Kishi-Kaboshi M, Aida R, Sasaki K. Parsley ubiquitin promoter displays higher activity than the CaMV 35S promoter and the Chrysanthemum actin 2 promoter for productive, constitutive, and durable expression of a transgene in Chrysanthemum morifolium. Breed Sci 2019;69:536-44. [CrossRef]
31. Poudel M. Functional Characterization of Putative Disease Resistance Genes of Strawberry Internet. Norwegian University of Life Sciences, Ås;2021. Available from:https://hdl.handle.net/11250/2828207 [Last accessed on Aug 06].
32. Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J Cell Mol Biol 2014;79:348-59. [CrossRef]
33. Sun X, Hu Z, Chen R, Jiang Q, Song G, Zhang H, et al. Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep 2015;5:10342. [CrossRef]
34. Wilson FM, Harrison K, Armitage AD, Simkin AJ, Harrison RJ. CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 2019;15:45. [CrossRef]