1. INTRODUCTION
Chitinases are enzymes that break down chitin into soluble and insoluble oligosaccharides of low molecular weight [1]. Chitin is a long-chain polymer of an N-acetylglucosamine, a glucose derivative. Chitin is known as a characteristic component of the cell walls of fungi, the exoskeletons of arthropods such as crustaceans and insects [2].
Because they produce a diversity of chitinases, many fungal species of Trichoderma genus are effective mycoparasites and are employed for biological control of fungal infections on agricultural crop plants [3]. However, certain chitinase genes from Trichoderma or other species such as rice and tobacco have been incorporated into various crops to aid them against fungal infections [4-10]. To date, there has been no record of chitinase gene transfer from Trichoderma, especially Trichoderma asperellum, into peanuts (Arachis hypogaea L.).
Peanut is a legume that is native to Central Brazil and is now widely grown worldwide, mainly for its edible oil and seeds. However, it is also one of the susceptible crops to serious diseases such as stem rot, root rot, and pod rot caused by numerous soil-borne pathogens such as Rhizoctonia solani, Aspergillus niger, and Sclerotium rolfsii [11-14].
The objective of this study was to generate pNHL20 plasmid, a plant expression vector containing the chitinase genes, and conjugate it in Agrobacterium tumefaciens LBA4404 to serve for gene transfer into peanuts in the next studies. Chitinase genes used for this aim include a wild-type gene Chi42 from T. asperellum SH16 and two synthetic genes syncodChi42-1 and syncodChi42-2 derived from chi42 have been optimized for the codon usage in plant expression from our previous study [15]. The chitinase genes are controlled by the peanut Asy root-specific promoter [16], and the chitinase enzyme will be secreted extracellularly by a signal peptide fused in front of the gene. An expression cassette designed as above promises to enable chitinase transgenic peanuts to resist soil-borne phytopathogenic fungi that cause root rot.
2. MATERIALS AND METHODS
2.1. Materials
The root-specific Asy promoter (NCBI: JQ780692) (562 bp long) was synthesized with two overhanging ends, HindIII and XbaI, and cloned in vector pUC19 by PHUSA Bio-Chemistry Co., Ltd. Plant expression binary pMYV719 vector [Figure 1] (about 12 kb long) was provided by Professor Yang Moon-Sik (Jeonbuk National University, Korea).
Genes encoding chitinase 42 kDa, including Chi42 (NCBI: HM191683.1), syncodChi42-1 (NCBI: MT083802.1), and syncodChi42-2 (NCBI: MT083803.1), with two XbaI and SacI overhang ends, and a signal peptide located at 5’ end (about 1.3 kb in total length) were also synthesized and cloned in pUC19 vector by PHUSA Bio-Chemistry Co., Ltd. Chi42 is a wild-type gene from T. asperellum SH16 [17], while syncodChi42-1 and syncodChi42-2 are synthetic genes generated from Chi42 that were optimized for codon usage for plant expression in our previous study [15].
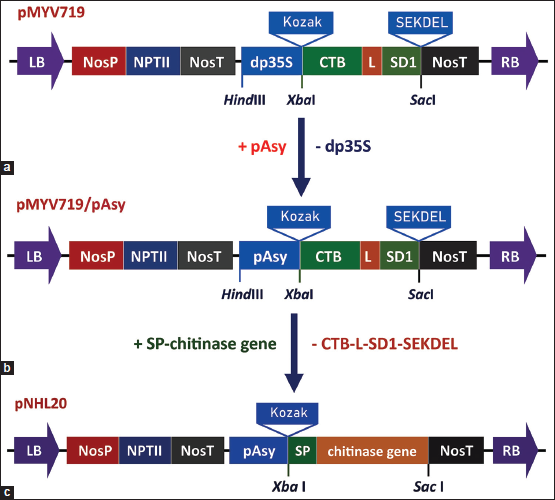 | Figure 1: Construction diagram of the plant expression vector pNHL20. (a) pMYV719 vector, (b) pMYV719/pAsy vector, and (c) pNHL20 vector. LB and RB: Left and right border of the T-DNA region, NosP: Promoter of nopaline synthase gene, NPTII: Neomycin phosphotransferase II gene for kanamycin resistance, NosT: Terminator of nopaline synthase gene, pAsy: Root-specific Asy promoter, Kozak: Consensus sequence to ribosomes recognize and initiate translation, CTB: Gene encoding cholera toxin B subunit, L: Sequence Gly-Pro-Gly-Pro, S1D: Epitope region S1D, SEKDEL: Maintenance signal in the endoplasmic reticulum, dp35S: Duplicated CaMV 35S promoter, SP: Signal peptide sequence, and chitinase gene: One of three genes Chi42, syncodChi42-1, and syncodChi42-2. [Click here to view] |
2.2. Construction of Plant Expression Binary Vector
The approach for constructing a plant expression vector is described in Figure 1. The Asy promoter (pAsy) was excised from the pUC19 vector using HindIII and XbaI and ligated into the pMYV719 vector at the location of the dp35S promoter (pdp35S), which was removed from the pMYV719 vector using the same enzymes, using T4 DNA ligase (Thermo Scientific).
XbaI and SacI were used to isolate chitinase genes from the pUC19 vector. Plant expression vectors pNHL20 were generated by removing CTB-L-SD1-SEKDEL sequences of around 1 kb from the recombinant pMYV719/pAsy vector with the same enzymes and replacing them with one of the chitinase genes.
The ligation reaction mixtures, pMYV719/pAsy and pNHL20 vectors, were transformed into Escherichia coli TOP10 (Thermo Scientific) by the heat-shock method [18]. The transformants were then selected on LB medium supplemented with 50 μg/mL kanamycin. The cloning efficiency was checked by restriction digestion of recombinant vector and electrophoresis on 0.8% agarose gel.
2.3. Triparental Mating
Three bacterial strains were conjugated (triparental mating) on LB medium, including A. tumefaciens LBA4404, E. coli TOP10 carrying the plant expression vector pNHL20, and E. coli TOP10 carrying the helper plasmid pRK2013. They were subsequently grown in the same medium but supplemented with 100 g/mL spectinomycin and 100 g/mL kanamycin. Screening for colonies of A. tumefaciens on the medium confirmed that the vectors pNHL20 had been conjugated [19].
2.4. Polymerase Chain Reaction (PCR) Amplification
The colony PCR method was used for transformed E. coli cells [20]. Plasmid DNA of A. tumefaciens LBA 4404 from triparental mating was prepared according to Kamble and Fawade [21].
PCR amplification was performed with specific primers of chitinase genes [Table 1]. The composition of the reaction consists of template DNA (20 ng of plasmid DNA from A. tumefaciens or E. coli colony), 10 pmol of each primer, 1 μL of Master Mix (Thermo Scientific), and water to a final volume of 12 μL. The PCR conditions were as follows: a genomic denaturation at 95°C for 15 min, followed by 30 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min. PCR products were then analyzed on 0.8% agarose gel.
 | Table 1: Nucleotide sequence of specific primers for chitinase genes [14]. [Click here to view] |
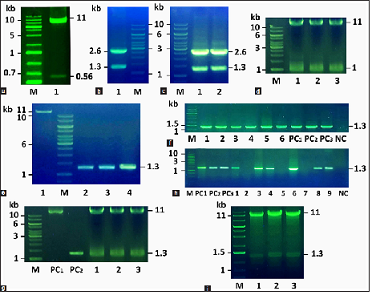 | Figure 2: Cloning of the Asy promoter and chitinase genes in plant expression vector pHNL20. (a) Restriction digestion of the recombinant pMYV719/pAsy vector by XbaI and HindIII. M: GeneRuler 1 kb Plus DNA Ladder (Thermo Scientific); 1: pMYV719 vector is about 11 kb long and pAsy promoter is about 0.56 kb long. (b and c) Restriction digestion of pUC19/chitinase vectors by XbaI and SacI. B1: chi42, C1: syncodChi42-1, and C2: syncodChi42-2. (d1–d3) Restriction digestion of pMYV719/Asy vectors by XbaI and SacI. (e) Purification of pMYV719/Asy vector and chitinase genes. 1: pMYV719/Asy vector and 2–4: Chi42, syncodChi42-1, and syncodChi42-2, respectively. (f) PCR amplification of chitinase genes with specific primers from transformed E. coli colonies. M: GeneRuler 1 kb DNA Ladder (Thermo Scientific), 1–2: Chi42, 3–4: syncodChi42-1, 5–6: syncodChi42–2, PC1: pUC19/Chi42 vector, PC2: pUC19/syncodChi42-1 vector, PC3: pUC19/syncodChi42-2 vector, and NC: Nontransformed E. coli. (g) Restriction digestion of derivatives of pNHL20 vector by XbaI và SacI. M: GeneRuler 1 kb DNA Ladder (Thermo Scientific), PC1: linearized pMYV719/Asy, PC2: Chi42 gene, and 1–3: pNHL20.1 for Chi42, pNHL20.2 for syncodChi42-1, and pNHL20.3 for syncodChi42-2. (h) PCR amplification of chitinase genes with specific primers from A. tumefaciens containing the derivatives of pNHL20 vector. M: GeneRuler 1 kb DNA Ladder (Thermo Scientific), 1–3: Chi42, 4–6: syncodChi42-1, 7–9: syncodChi42-2, PC1: pUC19/Chi42 vector, PC2: pUC19/syncodChi42-1 vector, PC3: pUC19/syncodChi42-2 vector, and NC: wild-type A. tumefaciens. (i) Restriction digestion of derivatives of pNHL20 vector by XbaI and SacI. M: GeneRuler 1 kb DNA Ladder (Thermo Scientific), 1: Chi42, 2: syncodChi42-1, and 3: syncodChi42-2. Chitinase gene is 1.3 kb long, and pNHL20 vector is 11 kb long. [Click here to view] |
3. RESULTS AND DISCUSSION
3.1. Vector pMYV719 Harboring pAsy
Promoter dp35S was removed from the pMYV719 vector using XbaI and HindIII. Promoter Asy was recovered from the pUC19/pAsy vector by the same two enzymes as above. The pMYV719 vector without the dp35S promoter was then ligated with the Asy promoter to transform into E. coli TOP10. The transformants were grown to produce the recombinant vector pMYV719/pAsy, which was then verified by restriction enzymes XbaI and HindIII. As a result, two DNA bands of approximately 11 kb and 560 bp were obtained, which corresponded to the vector and the pAsy sequence sizes [Figure 2a].
3.2. Vector pNHL20
Chitinase genes (Chi42, syncodChi42-1, and syncodChi42-2) with a signal peptide at the 5’ end were isolated from the pUC19/chitinase vectors to replace the CTB-L1-SD1-SEKDEL sequence in the pMYV719/pAsy plant expression vector. Figure 2b-d show restriction digestion of two vectors, pUC19/chitinase, and pMYV719/pAsy, using the same enzymes, XbaI and SacI. Chitinase genes are around 1.3 kb in length, while the pMYV719/pAsy vector lacking the CTB-L1-SD1-SEKDEL sequence is approximately 11 kb in length. The chitinase genes and the pMYV719/pAsy vector were purified with the GeneJET PCR Purification Kit (Thermo Fisher Scientific) before being bound to generate the vectors pNHL20.1/Chi42, pNHL20.2/syncodChi42-1, and pNHL20.3/syncodChi42-2 [Figure 2e]. The ligation mixture of pMYV719/pAsy vector and each chitinase gene was transformed into E. coli cells and the presence of chitinase genes was determined by PCR amplification and restriction digestion of putative pNHL20 vectors from colonies that grew in the selection medium [Figure 2f and g]. The current findings show plant expression vectors were successfully transformed into E. coli cells.
3.3. Triparental Mating
A. tumefaciens harboring variants of the pNHL20 plant expression vector, pNHL20.1, pNHL20.2, and pNHL20.3 were screened using PCR amplification and restriction digestion.
As shown in Figure 2h, PCR amplification revealed that these vectors were conjugated in some Agrobacterium colonies. Restriction digestion using XbaI and SacI confirmed these findings yet again; two DNA bands with predicted sizes of around 11 kb and 1.3 kb were detected on an agarose gel, which are compatible with pNHL20 vectors and chitinase genes, respectively [Figure 2i]. These findings indicate that A. tumefaciens carrying the binary vector is ready for chitinase gene transformation into peanuts in the subsequent investigation.
Promoters are a critical tool in biotechnological processes to ensure that the expression of a gene of interest is effective and well-controlled. When the interest protein is not required for the entire plant, the use of the organ or tissue-specific promoters that induce and specifically control the expression of transgenes in organs or tissue may be useful to prevent wasting energy and nutrients from transgenic plants [22]. Because peanuts are sensitive to various phytopathogenic fungi containing chitin in the cell wall, such as Rhizoctonia and Aspergillus, which cause root rot or collar rot, the production of extracellular chitinase in the roots is required to protect peanuts against these fungal pathogens. To date, there were no reports of the use of a root-specific promoter in peanuts yet, the majority of transgenic plants using the CaMV 35S constitutive promoters. The usage of constitutive promoters, however, results in unnecessary gene expression, potentially interfering with other plant growth pathways [23]. As a result, the goal of this work was to boost the antifungal effectiveness of peanuts by using a root-specific promoter.
As is known, microbial genes are generally weakly expressed in plants; therefore, the expression of heterologous proteins often requires modification of the coding sequence [24]. For that reason, the Chi42 gene encoding chitinase 42 kDa of fungus T. asperellum SH16 was optimized for codon usage for expression in plants to two new genes, syncodChi42-1 and syncodChi42-2, as in our previous report [15]. In the present study, each derivative of plant expression pNHL20 vector harboring one of three chitinase genes controlled by root-specific promoter was successfully introduced into A. tumefaciens LBA4404.
4. CONCLUSION
The chitinase genes (Chi42, syncodChi42-1, and syncodChi42-2) from T. asperellum SH16 with a signal peptide at 5’ end and root-specific Asy promoter were successfully cloned in plant expression vector pNHL20. This vector was transferred into A. tumefaciens LBA 4404 to serve the genetic transformation into peanuts for resistance to root rot disease caused by phytopathogenic fungi in further studies.
5. ACKNOWLEDGMENTS
The authors would also like to thank Hue University, Vietnam, for facilitating this study and Professor Yang Moon-Sik (Jeonbuk National University, South Korea) for providing the pMYV719 vector. Nguyen Hoang Tue and Phung Thi Bich Hoa were awarded scholarships from the Master/PhD Scholarship Program of Vingroup Innovation Fund (VINIF) and Vingroup Big Data Institute (VINBIGDATA) with corresponding codes VINIF.2021.ThS.46 and VINIF.2020.TS.111.
6. AUTHOR CONTRIBUTIONS
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work. All the authors are eligible to be an author as per the International Committee of Medical Journal Editors requirements/guidelines.
7. CONFLICTS OF INTEREST
The authors report no financial or any other conflicts of interest in this work.
8. ETHICAL APPROVALS
Not applicable.
9. FUNDING SOURCES
This work was supported by National Foundation for Science and Technology Development, Vietnam (Grant number 106.02-2017.346).
10. PUBLISHER’S NOTE
This journal remains neutral with regard to jurisdictional claims in published institutional affiliation.
REFERENCES
1. Kramer KJ, Muthukrishnan S. Insect chitinases: Molecular biology and potential use as biopesticides. Insect Biochem Mol Biol 1997;27:887-900. CrossRef
2. Tang WJ, Fernandez JG, Sohn JJ, Amemiya CT. Chitin is endogenously produced in vertebrates. Curr Biol 2015;25:897-900. CrossRef
3. Ihrmark K, Asmail N, Ubhayasekera W, Melin P, Stenlid J, Karlsson M. Comparative molecular evolution of trichoderma chitinases in response to mycoparasitic interactions. Evol Bioinform Online 2010;6:1-26. CrossRef
4. Nishizawa Y, Nishio Z, Nakazono K, Soma M, Nakajima E, Ugaki M, et al. Enhanced resistance to blast (Magnaporthe grisea) in transgenic Japonica rice by constitutive expression of rice chitinase. Theor Appl Genet 1999;99:383-90. CrossRef
5. Takahashi W, Fujimori M, Miura Y, Komatsu T, Nishizawa Y, Hibi T, et al. Increased resistance to crown rust disease in transgenic Italian ryegrass (Lolium multiflorum Lam.) expressing the rice chitinase gene. Plant Cell Rep 2005;23:811-8. CrossRef
6. Gentile A, Deng Z, La Malfa S, Distefano G, Domina F, Vitale A, et al. Enhanced resistance to Phoma tracheiphila and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed 2007;126:146-51. CrossRef
7. Baranski R, Klocke E, Nothnagel T. Chitinase CHIT36 from Trichoderma harzianum enhances resistance of transgenic carrot to fungal pathogens. J Phytopathol 2008;156:513-21. CrossRef
8. Zarinpanjeh N, Motallebi M, Zamani MR, Ziaei M. Enhanced resistance to Sclerotinia sclerotiorum in Brassica napus by co-expression of defensin and chimeric chitinase genes. J Appl Genet 2016;57:417-25. CrossRef
9. Ojaghian S, Wang L, Xie GL, Zhang JZ. Increased resistance against storage rot in transgenic carrots expressing chitinase chit42 from Trichoderma harzianum. Sci Hortic. 2018;234:81-6. CrossRef
10. Ojaghian S, Wang L, Xie GL, Zhang JZ. Effect of introducing chitinase gene on the resistance of tuber mustard against white. Plant Pathol J 2020;36:378-83. CrossRef
11. Ismail FM, Abd El-Momen SM. Effect of some soil amendments on yield and disease incidence in peanut (Arachis hypogaea L.). Egypt J Agric Res 2007;85:379-99.
12. Gour HN, Sharma P, Kaushal R. Pathological Aspects and Management of Root Rot of Groundnut: Root Rot of Groundnut (Sclerotium rolfsii Sacc.). Lap Lambert Academic Publishing; 2012.
13. Thiessen LD, Woodward JE. Diseases of peanut caused by soilborne pathogens in the Southwestern United States. Int Sch Res Notices 2012;2012:517905. CrossRef
14. Xu ML, Yang JG, Wu JX, Chi YC, Xie LH. First report of Aspergillus niger causing root rot of peanut in China. Plant Dis 2015;99:284. CrossRef
15. Luong NN, Tien NQ, Huy NX, Man LQ, Sinh DD, Tue NH, et al. Expression of 42 kDa chitinase (Ta-CHI42) of Trichoderma asperellum from a synthetic gene in Escherichia coli. FEMS Microbiol Lett 2021;368:fnab110. CrossRef
16. Geng L, Duan X, Liang C, Shu C, Song F, Zhang J. Mining tissue-specific contigs from peanut (Arachis hypogaea L.) for promoter cloning by deep transcriptome sequencing. Plant Cell Physiol 2014;55:1793-801. CrossRef
17. Loc NH, Quang HT, Hung NB, Huy ND, Phuong TT, Ha TT. Trichoderma asperellum Chi42 genes encode chitinase. Mycobiology 2011;39:182-6. CrossRef
18. Froger A, Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 2007;6:253. CrossRef
19. Tien NQ, Hoa PT, Tue NH, Thanh DV, Thi HA, Luong NN, et al. Transient expression of chi42 genes from Trichoderma asperellum in Nicotiana benthamiana by agroinfiltration. Int J Agric Biol 2021;26:177-84. CrossRef
20. Bergkessel M, Guthrie C. Colony PC. Methods Enzymol 2013;529:299-309. CrossRef
21. Kamble SP, Fawade MM. A rapid and inexpensive one-tube genomic DNA extraction method from Agrobacterium tumefaciens. 3 Biotech 2014;4:213-5. CrossRef
22. Grunennvaldt RL, Degenhardt-Goldbach J. Promoters used in genetic transformation of plants. Res J Biol Sci 2015;10:1-9.
23. De Paoli LG, Camargo RL, Harakava R, Mendes BM, Filho FA. Genetic transformation of “Valencia” orange with the cecropin MB39 gene. Braz Agric Res 2008;42:1663-6. CrossRef
24. Quax TE, Claassens NJ, Soll D, van der Oost J. Codon bias as a means to fine-tune gene expression. Mol Cell 2015;59:149-61. CrossRef